[English] 日本語
 Yorodumi
Yorodumi- EMDB-28625: Cryo-EM structure of cGMP bound truncated human CNGA3/CNGB3 chann... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of cGMP bound truncated human CNGA3/CNGB3 channel in lipid nanodisc, pre-open state | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | CNGA3 / CNGB3 / ligand-bound / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationinorganic cation import across plasma membrane / intracellular cyclic nucleotide activated cation channel complex / intracellularly cGMP-activated cation channel activity / intracellularly cAMP-activated cation channel activity / transmembrane transporter complex / axon initial segment / ligand-gated monoatomic ion channel activity / myosin binding / sodium channel activity / monoatomic cation transmembrane transport ...inorganic cation import across plasma membrane / intracellular cyclic nucleotide activated cation channel complex / intracellularly cGMP-activated cation channel activity / intracellularly cAMP-activated cation channel activity / transmembrane transporter complex / axon initial segment / ligand-gated monoatomic ion channel activity / myosin binding / sodium channel activity / monoatomic cation transmembrane transport / cGMP binding / response to magnesium ion / monoatomic cation transport / glial cell projection / photoreceptor outer segment / response to cAMP / visual perception / calcium channel activity / perikaryon / cadherin binding / dendrite / protein-containing complex binding / signal transduction / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.6 Å | |||||||||
 Authors Authors | Hu Z / Yang J | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Conformational trajectory of allosteric gating of the human cone photoreceptor cyclic nucleotide-gated channel. Authors: Zhengshan Hu / Xiangdong Zheng / Jian Yang /  Abstract: Cyclic nucleotide-gated (CNG) channels transduce chemical signals into electrical signals in sensory receptors and neurons. They are activated by cGMP or cAMP, which bind to the cyclic nucleotide- ...Cyclic nucleotide-gated (CNG) channels transduce chemical signals into electrical signals in sensory receptors and neurons. They are activated by cGMP or cAMP, which bind to the cyclic nucleotide-binding domain (CNBD) to open a gate located 50-60 Å away in the central cavity. Structures of closed and open vertebrate CNG channels have been solved, but the conformational landscape of this allosteric gating remains to be elucidated and enriched. Here, we report structures of the cGMP-activated human cone photoreceptor CNGA3/CNGB3 channel in closed, intermediate, pre-open and open states in detergent or lipid nanodisc, all with fully bound cGMP. The pre-open and open states are obtained only in the lipid nanodisc, suggesting a critical role of lipids in tuning the energetic landscape of CNGA3/CNGB3 activation. The different states exhibit subunit-unique, incremental and distinct conformational rearrangements that originate in the CNBD, propagate through the gating ring to the transmembrane domain, and gradually open the S6 cavity gate. Our work illustrates a spatial conformational-change wave of allosteric gating of a vertebrate CNG channel by its natural ligand and provides an expanded framework for studying CNG properties and channelopathy. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28625.map.gz emd_28625.map.gz | 109.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28625-v30.xml emd-28625-v30.xml emd-28625.xml emd-28625.xml | 17.2 KB 17.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_28625_fsc.xml emd_28625_fsc.xml | 10.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_28625.png emd_28625.png | 154.7 KB | ||
| Filedesc metadata |  emd-28625.cif.gz emd-28625.cif.gz | 6.2 KB | ||
| Others |  emd_28625_half_map_1.map.gz emd_28625_half_map_1.map.gz emd_28625_half_map_2.map.gz emd_28625_half_map_2.map.gz | 107.6 MB 107.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28625 http://ftp.pdbj.org/pub/emdb/structures/EMD-28625 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28625 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28625 | HTTPS FTP |
-Validation report
| Summary document |  emd_28625_validation.pdf.gz emd_28625_validation.pdf.gz | 1.2 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_28625_full_validation.pdf.gz emd_28625_full_validation.pdf.gz | 1.2 MB | Display | |
| Data in XML |  emd_28625_validation.xml.gz emd_28625_validation.xml.gz | 19 KB | Display | |
| Data in CIF |  emd_28625_validation.cif.gz emd_28625_validation.cif.gz | 24.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28625 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28625 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28625 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28625 | HTTPS FTP |
-Related structure data
| Related structure data |  8evbMC  8etpC  8eu3C  8eucC  8ev8C  8ev9C  8evaC  8evcC  28620  28621 C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_28625.map.gz / Format: CCP4 / Size: 115.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28625.map.gz / Format: CCP4 / Size: 115.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_28625_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
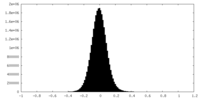
| Density Histograms |
-Half map: #1
| File | emd_28625_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
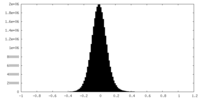
| Density Histograms |
- Sample components
Sample components
-Entire : Truncated human cone photoreceptor heterotetrameric CNG channel C...
| Entire | Name: Truncated human cone photoreceptor heterotetrameric CNG channel CNGA3/CNGB3 in complex with cGMP in lipid nanodisc |
|---|---|
| Components |
|
-Supramolecule #1: Truncated human cone photoreceptor heterotetrameric CNG channel C...
| Supramolecule | Name: Truncated human cone photoreceptor heterotetrameric CNG channel CNGA3/CNGB3 in complex with cGMP in lipid nanodisc type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Cyclic nucleotide-gated cation channel alpha-3
| Macromolecule | Name: Cyclic nucleotide-gated cation channel alpha-3 / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 63.402836 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MDYKDDDDKK TKKKDAIVVD PSSNLYYRWL TAIALPVFYN WYLLICRACF DELQSEYLML WLVLDYSADV LYVLDVLVRA RTGFLEQGL MVSDTNRLWQ HYKTTTQFKL DVLSLVPTDL AYLKVGTNYP EVRFNRLLKF SRLFEFFDRT ETRTNYPNMF R IGNLVLYI ...String: MDYKDDDDKK TKKKDAIVVD PSSNLYYRWL TAIALPVFYN WYLLICRACF DELQSEYLML WLVLDYSADV LYVLDVLVRA RTGFLEQGL MVSDTNRLWQ HYKTTTQFKL DVLSLVPTDL AYLKVGTNYP EVRFNRLLKF SRLFEFFDRT ETRTNYPNMF R IGNLVLYI LIIIHWNACI YFAISKFIGF GTDSWVYPNI SIPEHGRLSR KYIYSLYWST LTLTTIGETP PPVKDEEYLF VV VDFLVGV LIFATIVGNV GSMISNMNAS RAEFQAKIDS IKQYMQFRKV TKDLETRVIR WFDYLWANKK TVDEKEVLKS LPD KLKAEI AINVHLDTLK KVRIFQDCEA GLLVELVLKL RPTVFSPGDY ICKKGDIGKE MYIINEGKLA VVADDGVTQF VVLS DGSYF GEISILNIKG SKSGNRRTAN IRSIGYSDLF CLSKDDLMEA LTEYPEAKKA LEEKGRQILM KDNLIDEELA RAGAD PKDL EEKVEQLGSS LDTLQTRFAR LLAEYNATQM KMKQRLSQLE SQVKGGGDKP LADGEVPGDA TKTEDKQQ UniProtKB: Cyclic nucleotide-gated channel alpha-3 |
-Macromolecule #2: Cyclic nucleotide-gated cation channel beta-3
| Macromolecule | Name: Cyclic nucleotide-gated cation channel beta-3 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 84.637156 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MDYKDDDDKS GDLTTNPDPQ NAAEPTGTVP EQKEMDPGKE GPNSPQNKPP AAPVINEYAD AQLHNLVKRM RQRTALYKKK LVEGDLSSP EASPQTAKPT AVPPVKESDD KPTEHYYRLL WFKVKKMPLT EYLKRIKLPN SIDSYTDRLY LLWLLLVTLA Y NWNCCFIP ...String: MDYKDDDDKS GDLTTNPDPQ NAAEPTGTVP EQKEMDPGKE GPNSPQNKPP AAPVINEYAD AQLHNLVKRM RQRTALYKKK LVEGDLSSP EASPQTAKPT AVPPVKESDD KPTEHYYRLL WFKVKKMPLT EYLKRIKLPN SIDSYTDRLY LLWLLLVTLA Y NWNCCFIP LRLVFPYQTA DNIHYWLIAD IICDIIYLYD MLFIQPRLQF VRGGDIIVDS NELRKHYRTS TKFQLDVASI IP FDICYLF FGFNPMFRAN RMLKYTSFFE FNHHLESIMD KAYIYRVIRT TGYLLFILHI NACVYYWASN YEGIGTTRWV YDG EGNEYL RCYYWAVRTL ITIGGLPEPQ TLFEIVFQLL NFFSGVFVFS SLIGQMRDVI GAATANQNYF RACMDDTIAY MNNY SIPKL VQKRVRTWYE YTWDSQRMLD ESDLLKTLPT TVQLALAIDV NFSIISKVDL FKGCDTQMIY DMLLRLKSVL YLPGD FVCK KGEIGKEMYI IKHGEVQVLG GPDGTKVLVT LKAGSVFGEI SLLAAGGGNR RTANVVAHGF ANLLTLDKKT LQEILV HYP DSERILMKKA RVLLKQKAKT AEATPPRKDL ALLFPPKEET PKLFKTLLGG TGKASLARLL KLKREQAAQK KENSEGG EE EGKENEDKQK ENEDKQKENE DKGKENEDKD KGREPEEKPL DRPECTASPI AVEEEPHSVR RTVLPRGTSR QSLIISMA P SAEGGEEVLT IEVKEKAKQ UniProtKB: Cyclic nucleotide-gated channel beta-3 |
-Macromolecule #3: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 3 / Number of copies: 3 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #4: CYCLIC GUANOSINE MONOPHOSPHATE
| Macromolecule | Name: CYCLIC GUANOSINE MONOPHOSPHATE / type: ligand / ID: 4 / Number of copies: 4 / Formula: PCG |
|---|---|
| Molecular weight | Theoretical: 345.205 Da |
| Chemical component information |  ChemComp-PCG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8.58 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 61.05 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: OTHER / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.9 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)