+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | NuA4 HEAT | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
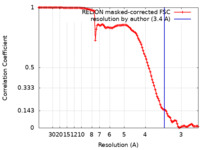
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Patel AB / Zukin SA / Nogales E | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2022 Journal: Elife / Year: 2022Title: Structure and flexibility of the yeast NuA4 histone acetyltransferase complex. Authors: Stefan A Zukin / Matthew R Marunde / Irina K Popova / Katarzyna M Soczek / Eva Nogales / Avinash B Patel /  Abstract: The NuA4 protein complex acetylates histones H4 and H2A to activate both transcription and DNA repair. We report the 3.1-Å resolution cryo-electron microscopy structure of the central hub of NuA4, ...The NuA4 protein complex acetylates histones H4 and H2A to activate both transcription and DNA repair. We report the 3.1-Å resolution cryo-electron microscopy structure of the central hub of NuA4, which flexibly tethers the histone acetyltransferase (HAT) and Trimer Independent of NuA4 involved in Transcription Interactions with Nucleosomes (TINTIN) modules. The hub contains the large Tra1 subunit and a core that includes Swc4, Arp4, Act1, Eaf1, and the C-terminal region of Epl1. Eaf1 stands out as the primary scaffolding factor that interacts with the Tra1, Swc4, and Epl1 subunits and contributes the conserved HSA helix to the Arp module. Using nucleosome-binding assays, we find that the HAT module, which is anchored to the core through Epl1, recognizes H3K4me3 nucleosomes with hyperacetylated H3 tails, while the TINTIN module, anchored to the core via Eaf1, recognizes nucleosomes that have hyperacetylated H2A and H4 tails. Together with the known interaction of Tra1 with site-specific transcription factors, our data suggest a model in which Tra1 recruits NuA4 to specific genomic sites then allowing the flexible HAT and TINTIN modules to select nearby nucleosomes for acetylation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28566.map.gz emd_28566.map.gz | 7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28566-v30.xml emd-28566-v30.xml emd-28566.xml emd-28566.xml | 13 KB 13 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_28566_fsc.xml emd_28566_fsc.xml | 11.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_28566.png emd_28566.png | 63.4 KB | ||
| Masks |  emd_28566_msk_1.map emd_28566_msk_1.map | 125 MB |  Mask map Mask map | |
| Others |  emd_28566_half_map_1.map.gz emd_28566_half_map_1.map.gz emd_28566_half_map_2.map.gz emd_28566_half_map_2.map.gz | 80.4 MB 80.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28566 http://ftp.pdbj.org/pub/emdb/structures/EMD-28566 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28566 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28566 | HTTPS FTP |
-Validation report
| Summary document |  emd_28566_validation.pdf.gz emd_28566_validation.pdf.gz | 571.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_28566_full_validation.pdf.gz emd_28566_full_validation.pdf.gz | 570.6 KB | Display | |
| Data in XML |  emd_28566_validation.xml.gz emd_28566_validation.xml.gz | 18.6 KB | Display | |
| Data in CIF |  emd_28566_validation.cif.gz emd_28566_validation.cif.gz | 24.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28566 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28566 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28566 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28566 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_28566.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28566.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.3404 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_28566_msk_1.map emd_28566_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_28566_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_28566_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : NuA4
| Entire | Name: NuA4 |
|---|---|
| Components |
|
-Supramolecule #1: NuA4
| Supramolecule | Name: NuA4 / type: complex / Chimera: Yes / ID: 1 / Parent: 0 / Macromolecule list: #1-#6 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 1.3 MDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.9 |
|---|---|
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: GRAPHENE OXIDE |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)