[English] 日本語
 Yorodumi
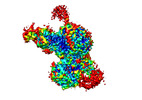
Yorodumi- EMDB-28547: Cryo-EM structure of PRC2 in complex with the long isoform of AEBP2 -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of PRC2 in complex with the long isoform of AEBP2 | ||||||||||||||||||||||||
 Map data Map data | |||||||||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||||||||
 Keywords Keywords | Polycomb repressive complex 2 Histone methyltransferase / GENE REGULATION | ||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationhepatocyte homeostasis / cellular response to trichostatin A / regulation of gliogenesis / negative regulation of striated muscle cell differentiation / regulation of kidney development / [histone H3]-lysine27 N-trimethyltransferase / response to tetrachloromethane / CAF-1 complex / histone H3K27 trimethyltransferase activity / negative regulation of keratinocyte differentiation ...hepatocyte homeostasis / cellular response to trichostatin A / regulation of gliogenesis / negative regulation of striated muscle cell differentiation / regulation of kidney development / [histone H3]-lysine27 N-trimethyltransferase / response to tetrachloromethane / CAF-1 complex / histone H3K27 trimethyltransferase activity / negative regulation of keratinocyte differentiation / negative regulation of retinoic acid receptor signaling pathway / cerebellar cortex development / primary miRNA binding / regulation of adaxial/abaxial pattern formation / random inactivation of X chromosome / skeletal muscle satellite cell maintenance involved in skeletal muscle regeneration / histone H3K27 methyltransferase activity / sex chromatin / regulatory ncRNA-mediated heterochromatin formation / positive regulation of cell cycle G1/S phase transition / NURF complex / facultative heterochromatin formation / NuRD complex / regulation of cell fate specification / negative regulation of stem cell population maintenance / genomic imprinting / DNA replication-dependent chromatin assembly / Transcription of E2F targets under negative control by p107 (RBL1) and p130 (RBL2) in complex with HDAC1 / ESC/E(Z) complex / negative regulation of stem cell differentiation / regulation of stem cell differentiation / protein-lysine N-methyltransferase activity / RSC-type complex / Polo-like kinase mediated events / cardiac muscle hypertrophy in response to stress / chromatin silencing complex / Transcription of E2F targets under negative control by DREAM complex / pronucleus / histone H3K9me2/3 reader activity / positive regulation of dendrite development / histone H3 methyltransferase activity / G1 to G0 transition / : / histone methyltransferase activity / ATPase complex / negative regulation of G1/S transition of mitotic cell cycle / DNA methylation-dependent constitutive heterochromatin formation / spinal cord development / negative regulation of gene expression, epigenetic / G1/S-Specific Transcription / lncRNA binding / synaptic transmission, GABAergic / histone deacetylase complex / Sin3-type complex / positive regulation of MAP kinase activity / Transcriptional Regulation by E2F6 / positive regulation of stem cell population maintenance / oligodendrocyte differentiation / RNA Polymerase I Transcription Initiation / negative regulation of transcription elongation by RNA polymerase II / G0 and Early G1 / positive regulation of protein serine/threonine kinase activity / positive regulation of GTPase activity / negative regulation of cell differentiation / positive regulation of epithelial to mesenchymal transition / ribonucleoprotein complex binding / subtelomeric heterochromatin formation / RNA polymerase II core promoter sequence-specific DNA binding / Cyclin E associated events during G1/S transition / pericentric heterochromatin / Transcriptional regulation of brown and beige adipocyte differentiation by EBF2 / Cyclin A:Cdk2-associated events at S phase entry / nucleosome binding / keratinocyte differentiation / Regulation of TP53 Activity through Acetylation / heterochromatin / protein localization to chromatin / negative regulation of cytokine production involved in inflammatory response / Deposition of new CENPA-containing nucleosomes at the centromere / liver regeneration / B cell differentiation / negative regulation of cell migration / SUMOylation of chromatin organization proteins / Regulation of PTEN gene transcription / transcription corepressor binding / stem cell differentiation / cellular response to leukemia inhibitory factor / ERCC6 (CSB) and EHMT2 (G9a) positively regulate rRNA expression / PRC2 methylates histones and DNA / Regulation of endogenous retroelements by KRAB-ZFP proteins / Defective pyroptosis / transcription coregulator activity / hippocampus development / Negative Regulation of CDH1 Gene Transcription / HDACs deacetylate histones / promoter-specific chromatin binding / negative regulation of transforming growth factor beta receptor signaling pathway / Regulation of endogenous retroelements by Piwi-interacting RNAs (piRNAs) / enzyme activator activity / G1/S transition of mitotic cell cycle Similarity search - Function | ||||||||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||||||||||||||
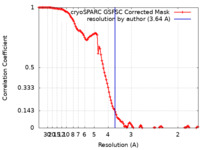
| Method | single particle reconstruction / cryo EM / Resolution: 3.64 Å | ||||||||||||||||||||||||
 Authors Authors | Boudes M / Zhang Q / Flanigan SF / Davidovich C | ||||||||||||||||||||||||
| Funding support |  Australia, 7 items Australia, 7 items
| ||||||||||||||||||||||||
 Citation Citation |  Journal: EMBO J / Year: 2025 Journal: EMBO J / Year: 2025Title: Auto-inhibition of PRC2 by the broadly expressed long isoform of AEBP2. Authors: Marlena Mucha / Zhihao Lai / Nicholas J McKenzie / Francesca Matrà / Marion Boudes / Sarena F Flanigan / Maria Teresa Alejo-Vinogradova / Craig Monger / Qi Zhang / Darragh Nimmo / Evan ...Authors: Marlena Mucha / Zhihao Lai / Nicholas J McKenzie / Francesca Matrà / Marion Boudes / Sarena F Flanigan / Maria Teresa Alejo-Vinogradova / Craig Monger / Qi Zhang / Darragh Nimmo / Evan Healy / Ademar J Silva / Daniel Angelov / David M Reck / Gráinne Holland / Zeynep Eda Atmaca / Helen E King / Maeve Hamilton / Eleanor Glancy / James Nolan / Robert J Weatheritt / Oliver Bell / Michiel Vermeulen / Chen Davidovich / Adrian P Bracken /     Abstract: Polycomb Repressive Complex 2 (PRC2) is an essential chromatin regulator responsible for mono-, di- and tri-methylating H3K27. Control of PRC2 activity is a critical process in development and ...Polycomb Repressive Complex 2 (PRC2) is an essential chromatin regulator responsible for mono-, di- and tri-methylating H3K27. Control of PRC2 activity is a critical process in development and disease, yet no inhibitory cofactor has been identified in somatic cells. Here, we show that the alternative isoforms of its accessory subunit AEBP2, namely AEBP2 (short) and AEBP2 (long), perform opposite functions in modulating PRC2 activity. Contrary to prior assumptions that AEBP2 enhances PRC2 function, we find that the widely expressed AEBP2 isoform inhibits it. AEBP2 is expressed throughout embryogenesis and adulthood and inhibits PRC2 DNA binding, histone methyltransferase activity, and binding to target genes. In contrast, AEBP2, expressed during early embryogenesis, promotes PRC2 DNA-binding activity and is essential for de novo repression of target genes during the transition from naïve to primed pluripotency. Mechanistically, through high-resolution cryo-EM and mutagenesis, we show that the recently evolved, negatively charged N-terminal region of AEBP2 inhibits PRC2. We propose a scenario in which the N-terminus of AEBP2 arose in vertebrates to restrain PRC2 activity in somatic cells. | ||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28547.map.gz emd_28547.map.gz | 88.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28547-v30.xml emd-28547-v30.xml emd-28547.xml emd-28547.xml | 33.3 KB 33.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_28547_fsc.xml emd_28547_fsc.xml | 12 KB | Display |  FSC data file FSC data file |
| Images |  emd_28547.png emd_28547.png | 73.3 KB | ||
| Filedesc metadata |  emd-28547.cif.gz emd-28547.cif.gz | 9.3 KB | ||
| Others |  emd_28547_additional_1.map.gz emd_28547_additional_1.map.gz emd_28547_half_map_1.map.gz emd_28547_half_map_1.map.gz emd_28547_half_map_2.map.gz emd_28547_half_map_2.map.gz | 168.1 MB 165.1 MB 165.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28547 http://ftp.pdbj.org/pub/emdb/structures/EMD-28547 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28547 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28547 | HTTPS FTP |
-Related structure data
| Related structure data |  8eqvMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_28547.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28547.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.86 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #1
| File | emd_28547_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_28547_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_28547_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Ternary complex of PRC2 with long isoform of AEBP2
| Entire | Name: Ternary complex of PRC2 with long isoform of AEBP2 |
|---|---|
| Components |
|
-Supramolecule #1: Ternary complex of PRC2 with long isoform of AEBP2
| Supramolecule | Name: Ternary complex of PRC2 with long isoform of AEBP2 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Histone-binding protein RBBP4
| Macromolecule | Name: Histone-binding protein RBBP4 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 47.709527 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MADKEAAFDD AVEERVINEE YKIWKKNTPF LYDLVMTHAL EWPSLTAQWL PDVTRPEGKD FSIHRLVLGT HTSDEQNHLV IASVQLPND DAQFDASHYD SEKGEFGGFG SVSGKIEIEI KINHEGEVNR ARYMPQNPCI IATKTPSSDV LVFDYTKHPS K PDPSGECN ...String: MADKEAAFDD AVEERVINEE YKIWKKNTPF LYDLVMTHAL EWPSLTAQWL PDVTRPEGKD FSIHRLVLGT HTSDEQNHLV IASVQLPND DAQFDASHYD SEKGEFGGFG SVSGKIEIEI KINHEGEVNR ARYMPQNPCI IATKTPSSDV LVFDYTKHPS K PDPSGECN PDLRLRGHQK EGYGLSWNPN LSGHLLSASD DHTICLWDIS AVPKEGKVVD AKTIFTGHTA VVEDVSWHLL HE SLFGSVA DDQKLMIWDT RSNNTSKPSH SVDAHTAEVN CLSFNPYSEF ILATGSADKT VALWDLRNLK LKLHSFESHK DEI FQVQWS PHNETILASS GTDRRLNVWD LSKIGEEQSP EDAEDGPPEL LFIHGGHTAK ISDFSWNPNE PWVICSVSED NIMQ VWQMA ENIYNDEDPE GSVDPEGQGS UniProtKB: Histone-binding protein RBBP4 |
-Macromolecule #2: Zinc finger protein AEBP2
| Macromolecule | Name: Zinc finger protein AEBP2 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 54.535496 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MAAAITDMAD LEELSRLSPL PPGSPGSAAR GRAEPPEEEE EEEEEEEEAE AEAVAALLLN GGSGGGGGGG GGGVGGGEAE TMSEPSPES ASQAGEDEDE EEDDEEEEDE SSSSGGGEEE SSAESLVGSS GGSSSDETRS LSPGAASSSS GDGDGKEGLE E PKGPRGSQ ...String: MAAAITDMAD LEELSRLSPL PPGSPGSAAR GRAEPPEEEE EEEEEEEEAE AEAVAALLLN GGSGGGGGGG GGGVGGGEAE TMSEPSPES ASQAGEDEDE EEDDEEEEDE SSSSGGGEEE SSAESLVGSS GGSSSDETRS LSPGAASSSS GDGDGKEGLE E PKGPRGSQ GGGGGGSSSS SVVSSGGDEG YGTGGGGSSA TSGGRRGSLE MSSDGEPLSR MDSEDSISST IMDVDSTISS GR STPAMMN GQGSTTSSSK NIAYNCCWDQ CQACFNSSPD LADHIRSIHV DGQRGGVFVC LWKGCKVYNT PSTSQSWLQR HML THSGDK PFKCVVGGCN ASFASQGGLA RHVPTHFSQQ NSSKVSSQPK AKEESPSKAG MNKRRKLKNK RRRSLPRPHD FFDA QTLDA IRHRAICFNL SAHIESLGKG HSVVFHSTVI AKRKEDSGKI KLLLHWMPED ILPDVWVNES ERHQLKTKVV HLSKL PKDT ALLLDPNIYR TMPQKRLKRT LIRKVFNLYL SKQ UniProtKB: Zinc finger protein AEBP2 |
-Macromolecule #3: Polycomb protein EED
| Macromolecule | Name: Polycomb protein EED / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 50.267691 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MSEREVSTAP AGTDMPAAKK QKLSSDENSN PDLSGDENDD AVSIESGTNT ERPDTPTNTP NAPGRKSWGK GKWKSKKCKY SFKCVNSLK EDHNQPLFGV QFNWHSKEGD PLVFATVGSN RVTLYECHSQ GEIRLLQSYV DADADENFYT CAWTYDSNTS H PLLAVAGS ...String: MSEREVSTAP AGTDMPAAKK QKLSSDENSN PDLSGDENDD AVSIESGTNT ERPDTPTNTP NAPGRKSWGK GKWKSKKCKY SFKCVNSLK EDHNQPLFGV QFNWHSKEGD PLVFATVGSN RVTLYECHSQ GEIRLLQSYV DADADENFYT CAWTYDSNTS H PLLAVAGS RGIIRIINPI TMQCIKHYVG HGNAINELKF HPRDPNLLLS VSKDHALRLW NIQTDTLVAI FGGVEGHRDE VL SADYDLL GEKIMSCGMD HSLKLWRINS KRMMNAIKES YDYNPNKTNR PFISQKIHFP DFSTRDIHRN YVDCVRWLGD LIL SKSCEN AIVCWKPGKM EDDIDKIKPS ESNVTILGRF DYSQCDIWYM RFSMDFWQKM LALGNQVGKL YVWDLEVEDP HKAK CTTLT HHKCGAAIRQ TSFSRDSSIL IAVCDDASIW RWDRLR UniProtKB: Polycomb protein EED |
-Macromolecule #4: Histone-lysine N-methyltransferase EZH2
| Macromolecule | Name: Histone-lysine N-methyltransferase EZH2 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO / EC number: [histone H3]-lysine27 N-trimethyltransferase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 85.492297 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MGQTGKKSEK GPVCWRKRVK SEYMRLRQLK RFRRADEVKS MFSSNRQKIL ERTEILNQEW KQRRIQPVHI LTSVSSLRGT RECSVTSDL DFPTQVIPLK TLNAVASVPI MYSWSPLQQN FMVEDETVLH NIPYMGDEVL DQDGTFIEEL IKNYDGKVHG D RECGFIND ...String: MGQTGKKSEK GPVCWRKRVK SEYMRLRQLK RFRRADEVKS MFSSNRQKIL ERTEILNQEW KQRRIQPVHI LTSVSSLRGT RECSVTSDL DFPTQVIPLK TLNAVASVPI MYSWSPLQQN FMVEDETVLH NIPYMGDEVL DQDGTFIEEL IKNYDGKVHG D RECGFIND EIFVELVNAL GQYNDDDDDD DGDDPEEREE KQKDLEDHRD DKESRPPRKF PSDKIFEAIS SMFPDKGTAE EL KEKYKEL TEQQLPGALP PECTPNIDGP NAKSVQREQS LHSFHTLFCR RCFKYDCFLH PFHATPNTYK RKNTETALDN KPC GPQCYQ HLEGAKEFAA ALTAERIKTP PKRPGGRRRG RLPNNSSRPS TPTINVLESK DTDSDREAGT ETGGENNDKE EEEK KDETS SSSEANSRCQ TPIKMKPNIE PPENVEWSGA EASMFRVLIG TYYDNFCAIA RLIGTKTCRQ VYEFRVKESS IIAPA PAED VDTPPRKKKR KHRLWAAHCR KIQLKKDGSS NHVYNYQPCD HPRQPCDSSC PCVIAQNFCE KFCQCSSECQ NRFPGC RCK AQCNTKQCPC YLAVRECDPD LCLTCGAADH WDSKNVSCKN CSIQRGSKKH LLLAPSDVAG WGIFIKDPVQ KNEFISE YC GEIISQDEAD RRGKVYDKYM CSFLFNLNND FVVDATRKGN KIRFANHSVN PNCYAKVMMV NGDHRIGIFA KRAIQTGE E LFFDYRYSQA DALKYVGIER EMEIP UniProtKB: Histone-lysine N-methyltransferase EZH2 |
-Macromolecule #5: Polycomb protein SUZ12
| Macromolecule | Name: Polycomb protein SUZ12 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 83.181922 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MAPQKHGGGG GGGSGPSAGS GGGGFGGSAA VAAATASGGK SGGGSCGGGG SYSASSSSSA AAAAGAAVLP VKKPKMEHVQ ADHELFLQA FEKPTQIYRF LRTRNLIAPI FLHRTLTYMS HRNSRTNIKR KTFKVDDMLS KVEKMKGEQE SHSLSAHLQL T FTGFFHKN ...String: MAPQKHGGGG GGGSGPSAGS GGGGFGGSAA VAAATASGGK SGGGSCGGGG SYSASSSSSA AAAAGAAVLP VKKPKMEHVQ ADHELFLQA FEKPTQIYRF LRTRNLIAPI FLHRTLTYMS HRNSRTNIKR KTFKVDDMLS KVEKMKGEQE SHSLSAHLQL T FTGFFHKN DKPSPNSENE QNSVTLEVLL VKVCHKKRKD VSCPIRQVPT GKKQVPLNPD LNQTKPGNFP SLAVSSNEFE PS NSHMVKS YSLLFRVTRP GRREFNGMIN GETNENIDVN EELPARRKRN REDGEKTFVA QMTVFDKNRR LQLLDGEYEV AMQ EMEECP ISKKRATWET ILDGKRLPPF ETFSQGPTLQ FTLRWTGETN DKSTAPIAKP LATRNSESLH QENKPGSVKP TQTI AVKES LTTDLQTRKE KDTPNENRQK LRIFYQFLYN NNTRQQTEAR DDLHCPWCTL NCRKLYSLLK HLKLCHSRFI FNYVY HPKG ARIDVSINEC YDGSYAGNPQ DIHRQPGFAF SRNGPVKRTP ITHILVCRPK RTKASMSEFL ESEDGEVEQQ RTYSSG HNR LYFHSDTCLP LRPQEMEVDS EDEKDPEWLR EKTITQIEEF SDVNEGEKEV MKLWNLHVMK HGFIADNQMN HACMLFV EN YGQKIIKKNL CRNFMLHLVS MHDFNLISIM SIDKAVTKLR EMQQKLEKGE SASPANEEIT EEQNGTANGF SEINSKEK A LETDSVSGVS KQSKKQKL UniProtKB: Polycomb protein SUZ12 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.4 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
Details: 200 mM NaCl, 20 mM HEPES pH 7.5, 1 mM TCEP, 0.01% NP-40 | |||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 120 sec. / Pretreatment - Atmosphere: AIR / Details: 0.24 mBar, 120 s, 10 mAmp | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV / Details: Blotting 3 seconds. | |||||||||||||||
| Details | Prior to cryo-EM sample preparation, complexes were PEGylated at 0.9 mg/mL with 5 mM MS(PEG)4 Methyl-PEG-NHS-Ester (ThermoFisher Scientific) for 2 h at 4oC. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum ER / Energy filter - Slit width: 10 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)