+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Type IIS Restriction Endonuclease PaqCI, DNA bound | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | homotetramer / restriction endonuclease / DNA binding / DNA cleavage / DNA BINDING PROTEIN | |||||||||
| Function / homology | Uncharacterized protein Function and homology information Function and homology information | |||||||||
| Biological species |  Paucibacter aquatile (bacteria) / DNA molecule (others) Paucibacter aquatile (bacteria) / DNA molecule (others) | |||||||||
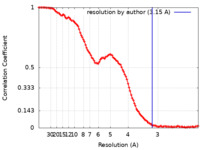
| Method | single particle reconstruction / cryo EM / Resolution: 3.15 Å | |||||||||
 Authors Authors | Kennedy MA / Stoddard BL | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nucleic Acids Res / Year: 2023 Journal: Nucleic Acids Res / Year: 2023Title: Structures, activity and mechanism of the Type IIS restriction endonuclease PaqCI. Authors: Madison A Kennedy / Christopher J Hosford / Caleigh M Azumaya / Yvette A Luyten / Minyong Chen / Richard D Morgan / Barry L Stoddard /  Abstract: Type IIS restriction endonucleases contain separate DNA recognition and catalytic domains and cleave their substrates at well-defined distances outside their target sequences. They are employed in ...Type IIS restriction endonucleases contain separate DNA recognition and catalytic domains and cleave their substrates at well-defined distances outside their target sequences. They are employed in biotechnology for a variety of purposes, including the creation of gene-targeting zinc finger and TAL effector nucleases and DNA synthesis applications such as Golden Gate assembly. The most thoroughly studied Type IIS enzyme, FokI, has been shown to require multimerization and engagement with multiple DNA targets for optimal cleavage activity; however, details of how it or similar enzymes forms a DNA-bound reaction complex have not been described at atomic resolution. Here we describe biochemical analyses of DNA cleavage by the Type IIS PaqCI restriction endonuclease and a series of molecular structures in the presence and absence of multiple bound DNA targets. The enzyme displays a similar tetrameric organization of target recognition domains in the absence or presence of bound substrate, with a significant repositioning of endonuclease domains in a trapped DNA-bound complex that is poised to deliver the first of a series of double-strand breaks. PaqCI and FokI share similar structural mechanisms of DNA cleavage, but considerable differences in their domain organization and quaternary architecture, facilitating comparisons between distinct Type IIS enzymes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28534.map.gz emd_28534.map.gz | 97.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28534-v30.xml emd-28534-v30.xml emd-28534.xml emd-28534.xml | 23.3 KB 23.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_28534_fsc.xml emd_28534_fsc.xml | 13.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_28534.png emd_28534.png | 101 KB | ||
| Masks |  emd_28534_msk_1.map emd_28534_msk_1.map | 103 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-28534.cif.gz emd-28534.cif.gz | 6.5 KB | ||
| Others |  emd_28534_half_map_1.map.gz emd_28534_half_map_1.map.gz emd_28534_half_map_2.map.gz emd_28534_half_map_2.map.gz | 95.7 MB 95.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28534 http://ftp.pdbj.org/pub/emdb/structures/EMD-28534 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28534 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28534 | HTTPS FTP |
-Related structure data
| Related structure data |  8epxMC  8em1C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_28534.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28534.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.122 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_28534_msk_1.map emd_28534_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map B
| File | emd_28534_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map A
| File | emd_28534_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : Homotetramer PaqCI with associated DNA duplexes
+Supramolecule #1: Homotetramer PaqCI with associated DNA duplexes
+Macromolecule #1: Type IIS Restriction Endonuclease PaqCI
+Macromolecule #2: DNA 4a
+Macromolecule #3: DNA 4b
+Macromolecule #4: DNA 2a
+Macromolecule #5: DNA 2b
+Macromolecule #6: DNA 3a
+Macromolecule #7: DNA 3b
+Macromolecule #8: DNA 1a
+Macromolecule #9: DNA 1b
+Macromolecule #10: CALCIUM ION
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.9 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 6.5 Component:
| ||||||||||||
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K | ||||||||||||
| Details | The DNA-bound PaqCI complex was generated by incubating enzyme + DNA at a 1:2 molar ratio. |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 1897 / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 36000 |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: OTHER / Overall B value: 139.1 |
|---|---|
| Output model |  PDB-8epx: |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)