+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | M. tuberculosis RNAP elongation complex with NusG | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Transcription elongation RNA polymerase pausing NusG cryo-EM / TRANSCRIPTION | |||||||||
| Function / homology |  Function and homology information Function and homology informationAntimicrobial action and antimicrobial resistance in Mtb / transcription elongation-coupled chromatin remodeling / bacterial-type RNA polymerase core enzyme binding / cytosolic DNA-directed RNA polymerase complex / DNA-directed RNA polymerase complex / peptidoglycan-based cell wall / regulation of DNA-templated transcription elongation / transcription antitermination / DNA-templated transcription initiation / DNA-templated transcription termination ...Antimicrobial action and antimicrobial resistance in Mtb / transcription elongation-coupled chromatin remodeling / bacterial-type RNA polymerase core enzyme binding / cytosolic DNA-directed RNA polymerase complex / DNA-directed RNA polymerase complex / peptidoglycan-based cell wall / regulation of DNA-templated transcription elongation / transcription antitermination / DNA-templated transcription initiation / DNA-templated transcription termination / ribonucleoside binding / DNA-directed RNA polymerase / DNA-directed RNA polymerase activity / protein dimerization activity / response to antibiotic / DNA-templated transcription / magnesium ion binding / DNA binding / zinc ion binding / plasma membrane / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Mycobacterium tuberculosis H37Rv (bacteria) / synthetic construct (others) Mycobacterium tuberculosis H37Rv (bacteria) / synthetic construct (others) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Vishwakarma RK / Murakami KS | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2023 Journal: Proc Natl Acad Sci U S A / Year: 2023Title: Allosteric mechanism of transcription inhibition by NusG-dependent pausing of RNA polymerase. Authors: Rishi K Vishwakarma / M Zuhaib Qayyum / Paul Babitzke / Katsuhiko S Murakami /  Abstract: NusG is a transcription elongation factor that stimulates transcription pausing in Gram+ bacteria including by sequence-specific interaction with a conserved pause-inducing TTNTTT motif found in the ...NusG is a transcription elongation factor that stimulates transcription pausing in Gram+ bacteria including by sequence-specific interaction with a conserved pause-inducing TTNTTT motif found in the non-template DNA (ntDNA) strand within the transcription bubble. To reveal the structural basis of NusG-dependent pausing, we determined a cryo-EM structure of a paused transcription complex (PTC) containing RNA polymerase (RNAP), NusG, and the TTNTTT motif in the ntDNA strand. The interaction of NusG with the ntDNA strand rearranges the transcription bubble by positioning three consecutive T residues in a cleft between NusG and the β-lobe domain of RNAP. We revealed that the RNAP swivel module rotation (swiveling), which widens (swiveled state) and narrows (non-swiveled state) a cleft between NusG and the β-lobe, is an intrinsic motion of RNAP and is directly linked to trigger loop (TL) folding, an essential conformational change of all cellular RNAPs for the RNA synthesis reaction. We also determined cryo-EM structures of RNAP escaping from the paused transcription state. These structures revealed the NusG-dependent pausing mechanism by which NusG-ntDNA interaction inhibits the transition from swiveled to non-swiveled states, thereby preventing TL folding and RNA synthesis allosterically. This motion is also reduced by the formation of an RNA hairpin within the RNA exit channel. Thus, the pause half-life can be modulated by the strength of the NusG-ntDNA interaction and/or the stability of the RNA hairpin. NusG residues that interact with the TTNTTT motif are widely conserved in bacteria, suggesting that NusG-dependent pausing is widespread. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28467.map.gz emd_28467.map.gz | 122 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28467-v30.xml emd-28467-v30.xml emd-28467.xml emd-28467.xml | 25.2 KB 25.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_28467.png emd_28467.png | 109.7 KB | ||
| Filedesc metadata |  emd-28467.cif.gz emd-28467.cif.gz | 8.5 KB | ||
| Others |  emd_28467_half_map_1.map.gz emd_28467_half_map_1.map.gz emd_28467_half_map_2.map.gz emd_28467_half_map_2.map.gz | 226.9 MB 226.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28467 http://ftp.pdbj.org/pub/emdb/structures/EMD-28467 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28467 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28467 | HTTPS FTP |
-Validation report
| Summary document |  emd_28467_validation.pdf.gz emd_28467_validation.pdf.gz | 957.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_28467_full_validation.pdf.gz emd_28467_full_validation.pdf.gz | 957.2 KB | Display | |
| Data in XML |  emd_28467_validation.xml.gz emd_28467_validation.xml.gz | 16.2 KB | Display | |
| Data in CIF |  emd_28467_validation.cif.gz emd_28467_validation.cif.gz | 19 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28467 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28467 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28467 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28467 | HTTPS FTP |
-Related structure data
| Related structure data |  8eotMC  8ehqC  8ej3C  8eoeC  8eofC  8eosC  8exyC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_28467.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28467.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.87 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_28467_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
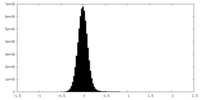
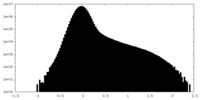
| Density Histograms |
-Half map: #1
| File | emd_28467_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
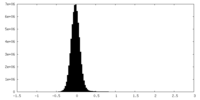
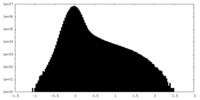
| Density Histograms |
- Sample components
Sample components
+Entire : Transcription elongation complex with M. tuberculosis NusG and CMPCPP
+Supramolecule #1: Transcription elongation complex with M. tuberculosis NusG and CMPCPP
+Macromolecule #1: DNA-directed RNA polymerase subunit alpha
+Macromolecule #2: DNA-directed RNA polymerase subunit beta'
+Macromolecule #3: DNA-directed RNA polymerase subunit omega
+Macromolecule #7: Transcription termination/antitermination protein NusG
+Macromolecule #8: DNA-directed RNA polymerase subunit beta
+Macromolecule #4: DNA (33-MER)
+Macromolecule #5: DNA (31-MER)
+Macromolecule #6: RNA (5'-R(P*AP*AP*GP*CP*GP*GP*AP*GP*AP*GP*GP*UP*A)-3')
+Macromolecule #9: MAGNESIUM ION
+Macromolecule #10: ZINC ION
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.9 |
|---|---|
| Grid | Model: Quantifoil R2/1 / Support film - Material: CARBON / Support film - topology: CONTINUOUS |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 283 K / Instrument: FEI VITROBOT MARK IV Details: Blot for 4-5 seconds before plunging in liquid ethane. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 45.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: OTHER / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.75 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.3 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 46900 |
| Initial angle assignment | Type: ANGULAR RECONSTITUTION |
| Final angle assignment | Type: PROJECTION MATCHING |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)