[English] 日本語
 Yorodumi
Yorodumi- EMDB-2845: 40S-eIF1-eIF1A-eIF3-eIF3j translation initiation complex from Lac... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-2845 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | 40S-eIF1-eIF1A-eIF3-eIF3j translation initiation complex from Lachancea kluyveri | |||||||||
 Map data Map data | Structure of the Lachancea kluyveri 40S-eIF1-eIF1A-eIF3-eIF3j initiation complex | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | eIF3 / translation initiation | |||||||||
| Function / homology |  Function and homology information Function and homology informationeukaryotic translation initiation factor 3 complex, eIF3e / eukaryotic translation initiation factor 3 complex, eIF3m / translation reinitiation / formation of cytoplasmic translation initiation complex / multi-eIF complex / eukaryotic translation initiation factor 3 complex / eukaryotic 43S preinitiation complex / eukaryotic 48S preinitiation complex / 90S preribosome / translation regulator activity ...eukaryotic translation initiation factor 3 complex, eIF3e / eukaryotic translation initiation factor 3 complex, eIF3m / translation reinitiation / formation of cytoplasmic translation initiation complex / multi-eIF complex / eukaryotic translation initiation factor 3 complex / eukaryotic 43S preinitiation complex / eukaryotic 48S preinitiation complex / 90S preribosome / translation regulator activity / translation initiation factor binding / translation initiation factor activity / maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / maturation of SSU-rRNA / small-subunit processome / rRNA processing / ribosome binding / ribosomal small subunit assembly / ribosomal small subunit biogenesis / small ribosomal subunit / small ribosomal subunit rRNA binding / cytosolic small ribosomal subunit / cytoplasmic translation / rRNA binding / structural constituent of ribosome / ribosome / translation / ribonucleoprotein complex / mRNA binding / nucleolus / RNA binding / zinc ion binding / metal ion binding / nucleus / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  Lachancea kluyveri (fungus) Lachancea kluyveri (fungus) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 6.47 Å | |||||||||
 Authors Authors | Aylett CHS / Boehringer D / Erzberger JP / Schaefer T / Ban N | |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2015 Journal: Nat Struct Mol Biol / Year: 2015Title: Structure of a yeast 40S-eIF1-eIF1A-eIF3-eIF3j initiation complex. Authors: Christopher H S Aylett / Daniel Boehringer / Jan P Erzberger / Tanja Schaefer / Nenad Ban /  Abstract: Eukaryotic translation initiation requires cooperative assembly of a large protein complex at the 40S ribosomal subunit. We have resolved a budding yeast initiation complex by cryo-EM, allowing ...Eukaryotic translation initiation requires cooperative assembly of a large protein complex at the 40S ribosomal subunit. We have resolved a budding yeast initiation complex by cryo-EM, allowing placement of prior structures of eIF1, eIF1A, eIF3a, eIF3b and eIF3c. Our structure highlights differences in initiation-complex binding to the ribosome compared to that of mammalian eIF3, demonstrates a direct contact between eIF3j and eIF1A and reveals the network of interactions between eIF3 subunits. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_2845.map.gz emd_2845.map.gz | 57.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-2845-v30.xml emd-2845-v30.xml emd-2845.xml emd-2845.xml | 21.2 KB 21.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emdb-2845.png emdb-2845.png | 127.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-2845 http://ftp.pdbj.org/pub/emdb/structures/EMD-2845 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2845 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2845 | HTTPS FTP |
-Related structure data
| Related structure data |  4uerMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_2845.map.gz / Format: CCP4 / Size: 62.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_2845.map.gz / Format: CCP4 / Size: 62.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Structure of the Lachancea kluyveri 40S-eIF1-eIF1A-eIF3-eIF3j initiation complex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.39 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
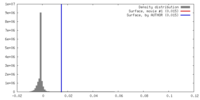
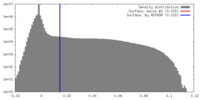
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 Movie
Movie Controller
Controller Sample components
Sample components


 UCSF Chimera
UCSF Chimera














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)




















 Processing
Processing Electron microscopy #1
Electron microscopy #1





