+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of E. coli CsgA fibril (260 pixel box size) | |||||||||
 Map data Map data | E coli CsgA fibril (260 pixel box size) sharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Curli / CsgA fibril / biofilm / PROTEIN FIBRIL | |||||||||
| Function / homology | Curlin associated / Curlin associated repeat / regulation of amyloid fibril formation / single-species biofilm formation / pilus / amyloid fibril formation / cell adhesion / identical protein binding / Major curlin subunit Function and homology information Function and homology information | |||||||||
| Biological species |  | |||||||||
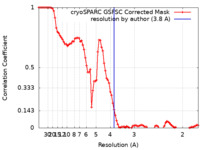
| Method | single particle reconstruction / cryo EM / Resolution: 3.8 Å | |||||||||
 Authors Authors | Bu F / Liu B | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: mBio / Year: 2024 Journal: mBio / Year: 2024Title: Structural insight into CsgA amyloid fibril assembly. Authors: Fan Bu / Derek R Dee / Bin Liu /   Abstract: The discovery of functional amyloids in bacteria dates back several decades, and our understanding of the curli biogenesis system has gradually expanded over time. However, due to its high ...The discovery of functional amyloids in bacteria dates back several decades, and our understanding of the curli biogenesis system has gradually expanded over time. However, due to its high aggregation propensity and intrinsically disordered nature, CsgA, the main structural component of curli fibrils, has eluded comprehensive structural characterization. Recent advancements in cryo-electron microscopy (cryo-EM) offer a promising tool to achieve high-resolution structural insights into CsgA fibrils. In this study, we outline an approach to addressing the colloidal instability challenges associated with CsgA, achieved through engineering and electrostatic repulsion. Then, we present the cryo-EM structure of CsgA fibrils at 3.62 Å resolution. This structure provides new insights into the cross-β structure of CsgA. Additionally, our study identifies two distinct spatial arrangements within several CsgA fibrils, a 2-CsgA-fibril pair and a 3-CsgA-fibril bundle, shedding light on the intricate hierarchy of the biofilm extracellular matrix and laying the foundation for precise manipulation of CsgA-derived biomaterials.IMPORTANCEThe visualization of the architecture of CsgA amyloid fibril has been a longstanding research question, for which a high-resolution structure is still unavailable. CsgA serves as a major subunit of curli, the primary component of the extracellular matrix generated by bacteria. The support provided by this extracellular matrix enables bacterial biofilms to resist antibiotic treatment, significantly impacting human health. CsgA has been identified in members of , with pathogenic being the most well-known model system. Our novel insights into the structure of CsgA protofilaments form the basis for drug design targeting diseases associated with biofilms. Additionally, CsgA is widely researched in biomaterials due to its self-assembly characteristics. The resolved spatial arrangements of CsgA amyloids revealed in our study will further enhance the precision design of functional biomaterials. Therefore, our study uniquely contributes to the understanding of CsgA amyloids for both microbiology and material science. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28277.map.gz emd_28277.map.gz | 63.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28277-v30.xml emd-28277-v30.xml emd-28277.xml emd-28277.xml | 18.3 KB 18.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_28277_fsc.xml emd_28277_fsc.xml | 8.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_28277.png emd_28277.png | 57.5 KB | ||
| Filedesc metadata |  emd-28277.cif.gz emd-28277.cif.gz | 5.5 KB | ||
| Others |  emd_28277_additional_1.map.gz emd_28277_additional_1.map.gz emd_28277_half_map_1.map.gz emd_28277_half_map_1.map.gz emd_28277_half_map_2.map.gz emd_28277_half_map_2.map.gz | 32.9 MB 62.2 MB 62.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28277 http://ftp.pdbj.org/pub/emdb/structures/EMD-28277 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28277 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28277 | HTTPS FTP |
-Related structure data
| Related structure data |  8enrMC  8enqC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_28277.map.gz / Format: CCP4 / Size: 67 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28277.map.gz / Format: CCP4 / Size: 67 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | E coli CsgA fibril (260 pixel box size) sharpened map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.88533 Å | ||||||||||||||||||||||||||||||||||||
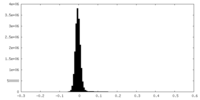
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: E coli CsgA fibril (260 pixel box size) unsharpened map
| File | emd_28277_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | E coli CsgA fibril (260 pixel box size) unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
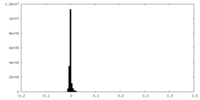
| Density Histograms |
-Half map: E coli CsgA fibril (260 pixel box size) half A map
| File | emd_28277_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | E coli CsgA fibril (260 pixel box size) half_A map | ||||||||||||
| Projections & Slices |
| ||||||||||||
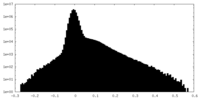
| Density Histograms |
-Half map: E coli CsgA fibril (260 pixel box size) half B map
| File | emd_28277_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | E coli CsgA fibril (260 pixel box size) half_B map | ||||||||||||
| Projections & Slices |
| ||||||||||||
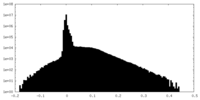
| Density Histograms |
- Sample components
Sample components
-Entire : E. coli CsgA fibril
| Entire | Name: E. coli CsgA fibril |
|---|---|
| Components |
|
-Supramolecule #1: E. coli CsgA fibril
| Supramolecule | Name: E. coli CsgA fibril / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Major curlin subunit
| Macromolecule | Name: Major curlin subunit / type: protein_or_peptide / ID: 1 / Number of copies: 8 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 14.095764 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGVVPQYGGG GNHGGGGNNS GPNSELNIYQ YGGGNSALAL QTDCRNSDLT ITQHGGGNGA DVGQGSDDSS IDLTQRGFGN SATLDQWNG KNSEMTVKQF GGGNGAAVDQ TASNSSVNVT QCGFGNNATA HQYHHHHHH UniProtKB: Major curlin subunit |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 10.4 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 295 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Number grids imaged: 1 / Number real images: 7550 / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.75 µm / Nominal magnification: 75300 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: OTHER |
|---|---|
| Output model |  PDB-8enr: |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)