[English] 日本語
 Yorodumi
Yorodumi- EMDB-28206: Clostridioides difficile binary toxin translocase CDTb wild-type ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Clostridioides difficile binary toxin translocase CDTb wild-type after calcium depletion from receptor binding domain 1 (RBD1) - Class 1 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | translocase / binary toxin / pore-forming / TOXIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationprotein homooligomerization / transferase activity / extracellular region / metal ion binding Similarity search - Function | |||||||||
| Biological species |  Clostridioides difficile (bacteria) Clostridioides difficile (bacteria) | |||||||||
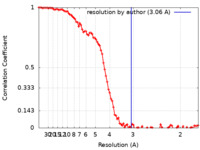
| Method | single particle reconstruction / cryo EM / Resolution: 3.06 Å | |||||||||
 Authors Authors | Abeyawardhane DL / Pozharski E | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Calcium-mediated Pore Formation of Clostridioides difficile Binary Toxin Authors: Abeyawardhane DL / Adipietro KA / Ruiz RG / Varney KM / Rustandi RR / Silin VI / Pozharski E / Weber DJ | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28206.map.gz emd_28206.map.gz | 32.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28206-v30.xml emd-28206-v30.xml emd-28206.xml emd-28206.xml | 20.1 KB 20.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_28206_fsc.xml emd_28206_fsc.xml | 11.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_28206.png emd_28206.png | 98.1 KB | ||
| Others |  emd_28206_half_map_1.map.gz emd_28206_half_map_1.map.gz emd_28206_half_map_2.map.gz emd_28206_half_map_2.map.gz | 59.5 MB 59.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28206 http://ftp.pdbj.org/pub/emdb/structures/EMD-28206 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28206 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28206 | HTTPS FTP |
-Related structure data
| Related structure data |  8eklMC  8ekkC  8ekmC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_28206.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28206.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.889 Å | ||||||||||||||||||||||||||||||||||||
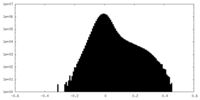
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_28206_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
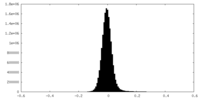
| Density Histograms |
-Half map: #2
| File | emd_28206_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
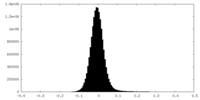
| Density Histograms |
- Sample components
Sample components
-Entire : Single heptamer (class 1) of Clostridioides difficile binary toxi...
| Entire | Name: Single heptamer (class 1) of Clostridioides difficile binary toxin translocase CDTb wild-type after depletion of calcium from RBD1 |
|---|---|
| Components |
|
-Supramolecule #1: Single heptamer (class 1) of Clostridioides difficile binary toxi...
| Supramolecule | Name: Single heptamer (class 1) of Clostridioides difficile binary toxin translocase CDTb wild-type after depletion of calcium from RBD1 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Clostridioides difficile (bacteria) / Strain: R20291 Clostridioides difficile (bacteria) / Strain: R20291 |
-Macromolecule #1: ADP-ribosyltransferase binding component
| Macromolecule | Name: ADP-ribosyltransferase binding component / type: protein_or_peptide / ID: 1 / Number of copies: 7 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Clostridioides difficile (bacteria) / Strain: R20291 Clostridioides difficile (bacteria) / Strain: R20291 |
| Molecular weight | Theoretical: 74.679086 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: LMSDWEDEDL DTDNDNIPDS YERNGYTIKD LIAVKWEDSF AEQGYKKYVS NYLESNTAGD PYTDYEKASG SFDKAIKTEA RDPLVAAYP IVGVGMEKLI ISTNEHASTD QGKTVSRATT NSKTESNTAG VSVNVGYQNG FTANVTTNYS HTTDNSTAVQ D SNGESWNT ...String: LMSDWEDEDL DTDNDNIPDS YERNGYTIKD LIAVKWEDSF AEQGYKKYVS NYLESNTAGD PYTDYEKASG SFDKAIKTEA RDPLVAAYP IVGVGMEKLI ISTNEHASTD QGKTVSRATT NSKTESNTAG VSVNVGYQNG FTANVTTNYS HTTDNSTAVQ D SNGESWNT GLSINKGESA YINANVRYYN TGTAPMYKVT PTTNLVLDGD TLSTIKAQEN QIGNNLSPGD TYPKKGLSPL AL NTMDQFS SRLIPINYDQ LKKLDAGKQI KLETTQVSGN FGTKNSSGQI VTEGNSWSDY ISQIDSISAS IILDTENESY ERR VTAKNL QDPEDKTPEL TIGEAIEKAF GATKKDGLLY FNDIPIDESC VELIFDDNTA NKIKDSLKTL SDKKIYNVKL ERGM NILIK TPTYFTNFDD YNNYPSTWSN VNTTNQDGLQ GSANKLNGET KIKIPMSELK PYKRYVFSGY SKDPLTSNSI IVKIK AKEE KTDYLVPEQG YTKFSYEFET TEKDSSNIEI TLIGSGTTYL DNLSITELNS TPEILDEPEV KIPTDQEIMD AHKIYF ADL NFNPSTGNTY INGMYFAPTQ TNKEALDYIQ KYRVEATLQY SGFKDIGTKD KEMRNYLGDP NQPKTNYVNL RSYFTGG EN IMTYKKLRIY AITPDDRELL VLSVD UniProtKB: ADP-ribosyltransferase binding component |
-Macromolecule #2: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 2 / Number of copies: 14 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)
































 Processing
Processing Electron microscopy
Electron microscopy FIELD EMISSION GUN
FIELD EMISSION GUN