+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Tail tip structure of Staphylococcus phage Andhra | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | phage tail / VIRUS | |||||||||
| Function / homology | CHAP domain profile. / CHAP domain / CHAP domain / Peptidase Function and homology information Function and homology information | |||||||||
| Biological species |  Staphylococcus phage Andhra (virus) Staphylococcus phage Andhra (virus) | |||||||||
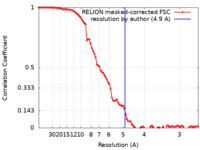
| Method | single particle reconstruction / cryo EM / Resolution: 4.9 Å | |||||||||
 Authors Authors | Kizziah JL / Hawkins NC / Dokland T | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2022 Journal: Sci Adv / Year: 2022Title: Structure and host specificity of bacteriophage Andhra. Authors: N'Toia C Hawkins / James L Kizziah / Asma Hatoum-Aslan / Terje Dokland /  Abstract: is an opportunistic pathogen of the human skin, often associated with infections of implanted medical devices. Staphylococcal picoviruses are a group of strictly lytic, short-tailed bacteriophages ... is an opportunistic pathogen of the human skin, often associated with infections of implanted medical devices. Staphylococcal picoviruses are a group of strictly lytic, short-tailed bacteriophages with compact genomes that are attractive candidates for therapeutic use. Here, we report the structure of the complete virion of -infecting phage Andhra, determined using high-resolution cryo-electron microscopy, allowing atomic modeling of 11 capsid and tail proteins. The capsid is a = 4 icosahedron containing a unique stabilizing capsid lining protein. The tail includes 12 trimers of a unique receptor binding protein (RBP), a lytic protein that also serves to anchor the RBPs to the tail stem, and a hexameric tail knob that acts as a gatekeeper for DNA ejection. Using structure prediction with AlphaFold, we identified the two proteins that comprise the tail tip heterooctamer. Our findings elucidate critical features for virion assembly, host recognition, and penetration. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28176.map.gz emd_28176.map.gz | 59.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28176-v30.xml emd-28176-v30.xml emd-28176.xml emd-28176.xml | 15.2 KB 15.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_28176_fsc.xml emd_28176_fsc.xml | 9.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_28176.png emd_28176.png | 77.5 KB | ||
| Filedesc metadata |  emd-28176.cif.gz emd-28176.cif.gz | 5.5 KB | ||
| Others |  emd_28176_half_map_1.map.gz emd_28176_half_map_1.map.gz emd_28176_half_map_2.map.gz emd_28176_half_map_2.map.gz | 49.8 MB 49.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28176 http://ftp.pdbj.org/pub/emdb/structures/EMD-28176 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28176 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28176 | HTTPS FTP |
-Related structure data
| Related structure data |  8ej5MC  8egrC  8egsC  8egtC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_28176.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28176.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.328 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_28176_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_28176_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Staphylococcus phage Andhra
| Entire | Name:  Staphylococcus phage Andhra (virus) Staphylococcus phage Andhra (virus) |
|---|---|
| Components |
|
-Supramolecule #1: Staphylococcus phage Andhra
| Supramolecule | Name: Staphylococcus phage Andhra / type: virus / ID: 1 / Parent: 0 / Macromolecule list: #2, #1 / NCBI-ID: 1958907 / Sci species name: Staphylococcus phage Andhra / Virus type: VIRION / Virus isolate: SPECIES / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  |
-Macromolecule #1: gp10, tail tip lysin (spike)
| Macromolecule | Name: gp10, tail tip lysin (spike) / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Staphylococcus phage Andhra (virus) Staphylococcus phage Andhra (virus) |
| Molecular weight | Theoretical: 51.675402 KDa |
| Recombinant expression | Organism:  Staphylococcus phage Andhra (virus) Staphylococcus phage Andhra (virus) |
| Sequence | String: MNDKEKIDKF IHSNLNDDFG LSVDDLVGKV KGIGRFSAWC GNSSTKIKQV LNAVKSIGVS PALFAAYEKN EGYNGSWGWL NHTSPQGNY LTDAQFVARK LVSQSRQAGT PSWIDAGNPV DFVPASVKRK GNYDFSHNMK NGKVGRAYIP LTAAATWAAY Y PEGLQASY ...String: MNDKEKIDKF IHSNLNDDFG LSVDDLVGKV KGIGRFSAWC GNSSTKIKQV LNAVKSIGVS PALFAAYEKN EGYNGSWGWL NHTSPQGNY LTDAQFVARK LVSQSRQAGT PSWIDAGNPV DFVPASVKRK GNYDFSHNMK NGKVGRAYIP LTAAATWAAY Y PEGLQASY NRVQNYGNPF LDAANTILGW GGKIDGKGGS SSGSSSSSSG TSGGLDVVAR AFEEFLKKLQ DSMQWDLHSI GT DKFFSNQ MFTITKTYNN TYRLNMNQKL LDEMKDLISR IDGGSGNDTG ADDSDGDHGG KAGKSVAPNG KSGRKIGGNW TYS NLPQKY KDAIEVPKFD PKYLAGSPFV NTGDTGQCTE LTWAYMHQIW GKRQPAWDNQ VTNGQRVWVV YRNQGARVTH RPTV GYGFS SKPNYLQAML PGVGHTGVVV AVFKDGSFLT ANYNVPPYWA PSRVVEYALI DGVPENAGDN IMFFSGIK UniProtKB: Peptidase |
-Macromolecule #2: gp1, tail tip protein
| Macromolecule | Name: gp1, tail tip protein / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Staphylococcus phage Andhra (virus) Staphylococcus phage Andhra (virus) |
| Molecular weight | Theoretical: 10.640743 KDa |
| Recombinant expression | Organism:  Staphylococcus phage Andhra (virus) Staphylococcus phage Andhra (virus) |
| Sequence | String: VTNEKGQAYT EMLQLFNLLQ QWNDFYTAEN ANNLLVACQQ LLINYNEPVI KFINDENEDK SLLQYLAGDD GLAQWQFYKG FYNNYNVHI F |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 39.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)