+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM reconstruction of the CFA/I bacterial adhesion pili | |||||||||
 Map data Map data | Primary map for the CFA/1 adhesion pili filament. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | enterotoxigenic / adhesion pili / superelastic / helical reconstruction / CELL ADHESION | |||||||||
| Function / homology | Fimbrial major subunit, CS1-type / CS1 type fimbrial major subunit / pilus / CFA/I fimbrial subunit B Function and homology information Function and homology information | |||||||||
| Biological species |  | |||||||||
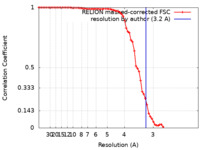
| Method | helical reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Doran MH / Bullitt E | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Structure / Year: 2023 Journal: Structure / Year: 2023Title: Three structural solutions for bacterial adhesion pilus stability and superelasticity. Authors: Matthew H Doran / Joseph L Baker / Tobias Dahlberg / Magnus Andersson / Esther Bullitt /   Abstract: Bacterial adhesion pili are key virulence factors that mediate host-pathogen interactions in diverse epithelial environments. Deploying a multimodal approach, we probed the structural basis ...Bacterial adhesion pili are key virulence factors that mediate host-pathogen interactions in diverse epithelial environments. Deploying a multimodal approach, we probed the structural basis underpinning the biophysical properties of pili originating from enterotoxigenic (ETEC) and uropathogenic bacteria. Using cryo-electron microscopy we solved the structures of three vaccine target pili from ETEC bacteria, CFA/I, CS17, and CS20. Pairing these and previous pilus structures with force spectroscopy and steered molecular dynamics simulations, we find a strong correlation between subunit-subunit interaction energies and the force required for pilus unwinding, irrespective of genetic similarity. Pili integrate three structural solutions for stabilizing their assemblies: layer-to-layer interactions, N-terminal interactions to distant subunits, and extended loop interactions from adjacent subunits. Tuning of these structural solutions alters the biophysical properties of pili and promotes the superelastic behavior that is essential for sustained bacterial attachment. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28150.map.gz emd_28150.map.gz | 28.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28150-v30.xml emd-28150-v30.xml emd-28150.xml emd-28150.xml | 16.1 KB 16.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_28150_fsc.xml emd_28150_fsc.xml | 7.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_28150.png emd_28150.png | 106.7 KB | ||
| Masks |  emd_28150_msk_1.map emd_28150_msk_1.map | 30.5 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-28150.cif.gz emd-28150.cif.gz | 5.6 KB | ||
| Others |  emd_28150_half_map_1.map.gz emd_28150_half_map_1.map.gz emd_28150_half_map_2.map.gz emd_28150_half_map_2.map.gz | 23.1 MB 23.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28150 http://ftp.pdbj.org/pub/emdb/structures/EMD-28150 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28150 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28150 | HTTPS FTP |
-Validation report
| Summary document |  emd_28150_validation.pdf.gz emd_28150_validation.pdf.gz | 921.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_28150_full_validation.pdf.gz emd_28150_full_validation.pdf.gz | 921.4 KB | Display | |
| Data in XML |  emd_28150_validation.xml.gz emd_28150_validation.xml.gz | 12.9 KB | Display | |
| Data in CIF |  emd_28150_validation.cif.gz emd_28150_validation.cif.gz | 17.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28150 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28150 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28150 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28150 | HTTPS FTP |
-Related structure data
| Related structure data |  8ehrMC  8ehsC  8ehtC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_28150.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28150.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Primary map for the CFA/1 adhesion pili filament. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.078 Å | ||||||||||||||||||||||||||||||||||||
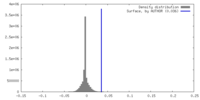
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_28150_msk_1.map emd_28150_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
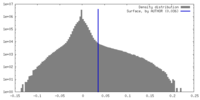
| Density Histograms |
-Half map: Half map 1.
| File | emd_28150_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1. | ||||||||||||
| Projections & Slices |
| ||||||||||||
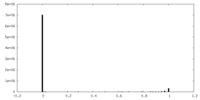
| Density Histograms |
-Half map: Half map 2.
| File | emd_28150_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2. | ||||||||||||
| Projections & Slices |
| ||||||||||||
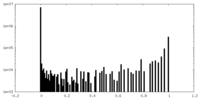
| Density Histograms |
- Sample components
Sample components
-Entire : CFA/I bacterial adhesion pili
| Entire | Name: CFA/I bacterial adhesion pili |
|---|---|
| Components |
|
-Supramolecule #1: CFA/I bacterial adhesion pili
| Supramolecule | Name: CFA/I bacterial adhesion pili / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: CFA/I fimbrial subunit B
| Macromolecule | Name: CFA/I fimbrial subunit B / type: protein_or_peptide / ID: 1 / Number of copies: 7 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 14.968839 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: VEKNITVTAS VDPAIDLLQA DGNALPSAVK LAYSPASKTF ESYRVMTQVH TNDATKKVIV KLADTPQLTD VLNSTVQMPI SVSWGGQVL STTAKEFEAA ALGYSASGVN GVSSSQELVI SAAPKTAGTA PTAGNYSGVV SLVMTLG UniProtKB: CFA/I fimbrial subunit B |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | helical array |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 400 / Support film - Material: GOLD / Support film - topology: HOLEY ARRAY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 15 sec. / Details: 15 mA with the Pelco EasiGlow Machine |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 283 K / Instrument: FEI VITROBOT MARK III |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number real images: 4809 / Average electron dose: 53.7 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)