+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Morphine-bound mu-opioid receptor-Gi complex | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | mu-opioid receptor / G protein / morphine / SIGNALING PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationOpioid Signalling / beta-endorphin receptor activity / morphine receptor activity / negative regulation of Wnt protein secretion / regulation of cellular response to stress / G protein-coupled opioid receptor signaling pathway / behavioral response to ethanol / negative regulation of nitric oxide biosynthetic process / sensory perception / adenylate cyclase-inhibiting G protein-coupled acetylcholine receptor signaling pathway ...Opioid Signalling / beta-endorphin receptor activity / morphine receptor activity / negative regulation of Wnt protein secretion / regulation of cellular response to stress / G protein-coupled opioid receptor signaling pathway / behavioral response to ethanol / negative regulation of nitric oxide biosynthetic process / sensory perception / adenylate cyclase-inhibiting G protein-coupled acetylcholine receptor signaling pathway / G-protein activation / Activation of the phototransduction cascade / Glucagon-type ligand receptors / Thromboxane signalling through TP receptor / Sensory perception of sweet, bitter, and umami (glutamate) taste / G beta:gamma signalling through PI3Kgamma / G beta:gamma signalling through CDC42 / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / Ca2+ pathway / G alpha (z) signalling events / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / regulation of NMDA receptor activity / Vasopressin regulates renal water homeostasis via Aquaporins / neuropeptide binding / Adrenaline,noradrenaline inhibits insulin secretion / ADP signalling through P2Y purinoceptor 12 / G alpha (q) signalling events / G alpha (i) signalling events / Thrombin signalling through proteinase activated receptors (PARs) / positive regulation of neurogenesis / Activation of G protein gated Potassium channels / G-protein activation / G beta:gamma signalling through PI3Kgamma / Prostacyclin signalling through prostacyclin receptor / G beta:gamma signalling through PLC beta / ADP signalling through P2Y purinoceptor 1 / Thromboxane signalling through TP receptor / Presynaptic function of Kainate receptors / G beta:gamma signalling through CDC42 / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G alpha (12/13) signalling events / Glucagon-type ligand receptors / G beta:gamma signalling through BTK / ADP signalling through P2Y purinoceptor 12 / Adrenaline,noradrenaline inhibits insulin secretion / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / Ca2+ pathway / Thrombin signalling through proteinase activated receptors (PARs) / G alpha (z) signalling events / Extra-nuclear estrogen signaling / G alpha (s) signalling events / photoreceptor outer segment membrane / G alpha (q) signalling events / negative regulation of cytosolic calcium ion concentration / spectrin binding / G alpha (i) signalling events / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / Vasopressin regulates renal water homeostasis via Aquaporins / alkylglycerophosphoethanolamine phosphodiesterase activity / G-protein alpha-subunit binding / photoreceptor outer segment / G protein-coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger / neuropeptide signaling pathway / voltage-gated calcium channel activity / adenylate cyclase inhibitor activity / MECP2 regulates neuronal receptors and channels / positive regulation of protein localization to cell cortex / T cell migration / Adenylate cyclase inhibitory pathway / D2 dopamine receptor binding / response to prostaglandin E / cardiac muscle cell apoptotic process / photoreceptor inner segment / G protein-coupled serotonin receptor binding / adenylate cyclase regulator activity / adenylate cyclase-inhibiting serotonin receptor signaling pathway / sensory perception of pain / cellular response to forskolin / Peptide ligand-binding receptors / regulation of mitotic spindle organization / Regulation of insulin secretion / positive regulation of cholesterol biosynthetic process / negative regulation of insulin secretion / G protein-coupled receptor binding / G protein-coupled receptor activity / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / response to peptide hormone / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / centriolar satellite / G-protein beta/gamma-subunit complex binding / G-protein activation / ADP signalling through P2Y purinoceptor 12 / GDP binding / Adrenaline,noradrenaline inhibits insulin secretion / G alpha (z) signalling events / cellular response to catecholamine stimulus Similarity search - Function | ||||||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /   | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | ||||||||||||
 Authors Authors | Zhuang Y / Wang Y / Guo S / Zhou XE / Rao Q / He X / He B / Liu J / Zhou Q / Wang X ...Zhuang Y / Wang Y / Guo S / Zhou XE / Rao Q / He X / He B / Liu J / Zhou Q / Wang X / Liu W / Jiang X / Yang D / Chen X / Jiang Y / Jiang H / Shen J / Melcher K / Wang M / Xie X / Xu HE | ||||||||||||
| Funding support |  China, 3 items China, 3 items
| ||||||||||||
 Citation Citation |  Journal: Cell / Year: 2022 Journal: Cell / Year: 2022Title: Molecular recognition of morphine and fentanyl by the human μ-opioid receptor. Authors: Youwen Zhuang / Yue Wang / Bingqing He / Xinheng He / X Edward Zhou / Shimeng Guo / Qidi Rao / Jiaqi Yang / Jinyu Liu / Qingtong Zhou / Xiaoxi Wang / Mingliang Liu / Weiyi Liu / Xiangrui ...Authors: Youwen Zhuang / Yue Wang / Bingqing He / Xinheng He / X Edward Zhou / Shimeng Guo / Qidi Rao / Jiaqi Yang / Jinyu Liu / Qingtong Zhou / Xiaoxi Wang / Mingliang Liu / Weiyi Liu / Xiangrui Jiang / Dehua Yang / Hualiang Jiang / Jingshan Shen / Karsten Melcher / Hong Chen / Yi Jiang / Xi Cheng / Ming-Wei Wang / Xin Xie / H Eric Xu /   Abstract: Morphine and fentanyl are among the most used opioid drugs that confer analgesia and unwanted side effects through both G protein and arrestin signaling pathways of μ-opioid receptor (μOR). Here, ...Morphine and fentanyl are among the most used opioid drugs that confer analgesia and unwanted side effects through both G protein and arrestin signaling pathways of μ-opioid receptor (μOR). Here, we report structures of the human μOR-G protein complexes bound to morphine and fentanyl, which uncover key differences in how they bind the receptor. We also report structures of μOR bound to TRV130, PZM21, and SR17018, which reveal preferential interactions of these agonists with TM3 side of the ligand-binding pocket rather than TM6/7 side. In contrast, morphine and fentanyl form dual interactions with both TM3 and TM6/7 regions. Mutations at the TM6/7 interface abolish arrestin recruitment of μOR promoted by morphine and fentanyl. Ligands designed to reduce TM6/7 interactions display preferential G protein signaling. Our results provide crucial insights into fentanyl recognition and signaling of μOR, which may facilitate rational design of next-generation analgesics. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28069.map.gz emd_28069.map.gz | 38 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28069-v30.xml emd-28069-v30.xml emd-28069.xml emd-28069.xml | 25.3 KB 25.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_28069.png emd_28069.png | 117.2 KB | ||
| Filedesc metadata |  emd-28069.cif.gz emd-28069.cif.gz | 7.5 KB | ||
| Others |  emd_28069_half_map_1.map.gz emd_28069_half_map_1.map.gz emd_28069_half_map_2.map.gz emd_28069_half_map_2.map.gz | 31.3 MB 31.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28069 http://ftp.pdbj.org/pub/emdb/structures/EMD-28069 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28069 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28069 | HTTPS FTP |
-Related structure data
| Related structure data |  8ef6MC  8ef5C  8efbC  8eflC  8efoC  8efqC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_28069.map.gz / Format: CCP4 / Size: 40.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28069.map.gz / Format: CCP4 / Size: 40.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.071 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_28069_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
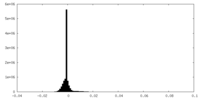
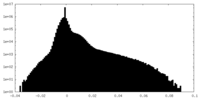
| Density Histograms |
-Half map: #2
| File | emd_28069_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
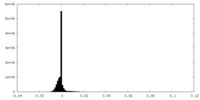
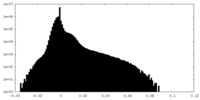
| Density Histograms |
- Sample components
Sample components
-Entire : fentanyl bound mu-opioid receptor-G protein complex
| Entire | Name: fentanyl bound mu-opioid receptor-G protein complex |
|---|---|
| Components |
|
-Supramolecule #1: fentanyl bound mu-opioid receptor-G protein complex
| Supramolecule | Name: fentanyl bound mu-opioid receptor-G protein complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Mu-type opioid receptor
| Macromolecule | Name: Mu-type opioid receptor / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 41.072191 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DSSAAPTNAS NCTDALAYSS CSPAPSPGSW VNLSHLDGNL SDPCGPNRTD LGGRDSLCPP TGSPSMITAI TIMALYSIVC VVGLFGNFL VMYVIVRYTK MKTATNIYIF NLALADALAT STLPFQSVNY LMGTWPFGTI LCKIVISIDY YNMFTSIFTL C TMSVDRYI ...String: DSSAAPTNAS NCTDALAYSS CSPAPSPGSW VNLSHLDGNL SDPCGPNRTD LGGRDSLCPP TGSPSMITAI TIMALYSIVC VVGLFGNFL VMYVIVRYTK MKTATNIYIF NLALADALAT STLPFQSVNY LMGTWPFGTI LCKIVISIDY YNMFTSIFTL C TMSVDRYI AVCHPVKALD FRTPRNAKII NVCNWILSSA IGLPVMFMAT TKYRQGSIDC TLTFSHPTWY WENLLKICVF IF AFIMPVL IITVCYGLMI LRLKSVRMLS GSKEKDRNLR RITRMVLVVV AVFIVCWTPI HIYVIIKALV TIPETTFQTV SWH FCIALG YTNSCLNPVL YAFLDENFKR CFREFCIPTS SNIEQQNSTR I UniProtKB: Mu-type opioid receptor |
-Macromolecule #2: Guanine nucleotide-binding protein G(i) subunit alpha-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(i) subunit alpha-1 type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 40.445059 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGCTLSAEDK AAVERSKMID RNLREDGEKA AREVKLLLLG AGESGKSTIV KQMKIIHEAG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI ...String: MGCTLSAEDK AAVERSKMID RNLREDGEKA AREVKLLLLG AGESGKSTIV KQMKIIHEAG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI PTQQDVLRTR VKTTGIVETH FTFKDLHFKM FDVGAQRSER KKWIHCFEGV TAIIFCVALS DYDLVLAEDE EM NRMHESM KLFDSICNNK WFTDTSIILF LNKKDLFEEK IKKSPLTICY PEYAGSNTYE EAAAYIQCQF EDLNKRKDTK EIY THFTCS TDTKNVQFVF DAVTDVIIKN NLKDCGLF UniProtKB: Guanine nucleotide-binding protein G(i) subunit alpha-1 |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 39.020664 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MHHHHHHHHG SLLQSELDQL RQEAEQLKNQ IRDARKACAD ATLSQITNNI DPVGRIQMRT RRTLRGHLAK IYAMHWGTDS RLLVSASQD GKLIIWDSYT TNKVHAIPLR SSWVMTCAYA PSGNYVACGG LDNICSIYNL KTREGNVRVS RELAGHTGYL S CCRFLDDN ...String: MHHHHHHHHG SLLQSELDQL RQEAEQLKNQ IRDARKACAD ATLSQITNNI DPVGRIQMRT RRTLRGHLAK IYAMHWGTDS RLLVSASQD GKLIIWDSYT TNKVHAIPLR SSWVMTCAYA PSGNYVACGG LDNICSIYNL KTREGNVRVS RELAGHTGYL S CCRFLDDN QIVTSSGDTT CALWDIETGQ QTTTFTGHTG DVMSLSLAPD TRLFVSGACD ASAKLWDVRE GMCRQTFTGH ES DINAICF FPNGNAFATG SDDATCRLFD LRADQELMTY SHDNIICGIT SVSFSKSGRL LLAGYDDFNC NVWDALKADR AGV LAGHDN RVSCLGVTDD GMAVATGSWD SFLKIWN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #4: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 7.56375 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFC UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #5: scFV16
| Macromolecule | Name: scFV16 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 26.408492 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MVQLVESGGG LVQPGGSRKL SCSASGFAFS SFGMHWVRQA PEKGLEWVAY ISSGSGTIYY ADTVKGRFTI SRDDPKNTLF LQMTSLRSE DTAMYYCVRS IYYYGSSPFD FWGQGTTLTV SAGGGGSGGG GSGGGGSADI VMTQATSSVP VTPGESVSIS C RSSKSLLH ...String: MVQLVESGGG LVQPGGSRKL SCSASGFAFS SFGMHWVRQA PEKGLEWVAY ISSGSGTIYY ADTVKGRFTI SRDDPKNTLF LQMTSLRSE DTAMYYCVRS IYYYGSSPFD FWGQGTTLTV SAGGGGSGGG GSGGGGSADI VMTQATSSVP VTPGESVSIS C RSSKSLLH SNGNTYLYWF LQRPGQSPQL LIYRMSNLAS GVPDRFSGSG SGTAFTLTIS RLEAEDVGVY YCMQHLEYPL TF GAGTKLE L |
-Macromolecule #6: (7R,7AS,12BS)-3-METHYL-2,3,4,4A,7,7A-HEXAHYDRO-1H-4,12-METHANO[1]...
| Macromolecule | Name: (7R,7AS,12BS)-3-METHYL-2,3,4,4A,7,7A-HEXAHYDRO-1H-4,12-METHANO[1]BENZOFURO[3,2-E]ISOQUINOLINE-7,9-DIOL type: ligand / ID: 6 / Number of copies: 2 / Formula: MOI |
|---|---|
| Molecular weight | Theoretical: 285.338 Da |
| Chemical component information | 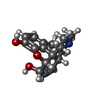 ChemComp-MOI: |
-Macromolecule #7: CHOLESTEROL
| Macromolecule | Name: CHOLESTEROL / type: ligand / ID: 7 / Number of copies: 10 / Formula: CLR |
|---|---|
| Molecular weight | Theoretical: 386.654 Da |
| Chemical component information |  ChemComp-CLR: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.2 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 70.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 5.0 µm / Nominal defocus min: 3.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)