+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of engineered nano-cage fusion protein | |||||||||
 Map data Map data | sharpened icosahedral map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | nano-cage / self-assembly / fusion protein / PROTEIN BINDING | |||||||||
| Function / homology | D-galactonate catabolic process / 2-dehydro-3-deoxy-6-phosphogalactonate aldolase activity / KDPG/KHG aldolase / KDPG and KHG aldolase / Aldolase-type TIM barrel / 2-dehydro-3-deoxyphosphogluconate aldolase/4-hydroxy-2-oxoglutarate aldolase Function and homology information Function and homology information | |||||||||
| Biological species |   Thermotoga maritima (bacteria) Thermotoga maritima (bacteria) | |||||||||
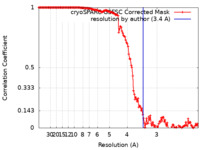
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Moustafa IM / Hafenstein SL | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: bioRxiv / Year: 2022 Journal: bioRxiv / Year: 2022Title: Intranasal SARS-CoV-2 RBD decorated nanoparticle vaccine enhances viral clearance in the Syrian hamster model. Authors: D R Patel / A M Minns / D G Sim / C J Field / A E Kerr / T Heinly / E H Luley / R M Rossi / C Bator / I M Moustafa / S L Hafenstein / S E Lindner / T C Sutton /  Abstract: Multiple vaccines have been developed and licensed for SARS-CoV-2. While these vaccines reduce disease severity, they do not prevent infection, and SARS-CoV-2 continues to spread and evolve. To ...Multiple vaccines have been developed and licensed for SARS-CoV-2. While these vaccines reduce disease severity, they do not prevent infection, and SARS-CoV-2 continues to spread and evolve. To prevent infection and limit transmission, vaccines must be developed that induce immunity in the respiratory tract. Therefore, we performed proof-of-principle vaccination studies with an intranasal nanoparticle vaccine against SARS-CoV-2. The vaccine candidate consisted of the self-assembling 60-subunit I3-01 protein scaffold covalently decorated with the SARS-CoV-2 receptor binding domain (RBD) using the SpyCatcher-SpyTag system. We verified the intended antigen display features by reconstructing the I3-01 scaffold to 3.4A using cryo-EM, and then demonstrated that the scaffold was highly saturated when grafted with RBD. Using this RBD-grafted SpyCage scaffold (RBD+SpyCage), we performed two unadjuvanted intranasal vaccination studies in the "gold-standard" preclinical Syrian hamster model. Hamsters received two vaccinations 28 days apart, and were then challenged 28 days post-boost with SARS-CoV-2. The initial study focused on assessing the immunogenicity of RBD+SpyCage, which indicated that vaccination of hamsters induced a non-neutralizing antibody response that enhanced viral clearance but did not prevent infection. In an expanded study, we demonstrated that covalent bonding of RBD to the scaffold was required to induce an antibody response. Consistent with the initial study, animals vaccinated with RBD+SpyCage more rapidly cleared SARS-CoV-2 from both the upper and lower respiratory tract. These findings demonstrate the intranasal SpyCage vaccine platform can induce protection against SARS-CoV-2 and, with additional modifications to improve immunogenicity, is a versatile platform for the development of intranasal vaccines targeting respiratory pathogens. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27812.map.gz emd_27812.map.gz | 266.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27812-v30.xml emd-27812-v30.xml emd-27812.xml emd-27812.xml | 17.5 KB 17.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_27812_fsc.xml emd_27812_fsc.xml | 13.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_27812.png emd_27812.png | 145.4 KB | ||
| Masks |  emd_27812_msk_1.map emd_27812_msk_1.map | 282.6 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-27812.cif.gz emd-27812.cif.gz | 6 KB | ||
| Others |  emd_27812_half_map_1.map.gz emd_27812_half_map_1.map.gz emd_27812_half_map_2.map.gz emd_27812_half_map_2.map.gz | 261.8 MB 261.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27812 http://ftp.pdbj.org/pub/emdb/structures/EMD-27812 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27812 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27812 | HTTPS FTP |
-Related structure data
| Related structure data |  8e01MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_27812.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27812.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened icosahedral map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_27812_msk_1.map emd_27812_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: halfB map
| File | emd_27812_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | halfB map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: halfA map
| File | emd_27812_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | halfA map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Nano-cage
| Entire | Name: Nano-cage |
|---|---|
| Components |
|
-Supramolecule #1: Nano-cage
| Supramolecule | Name: Nano-cage / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Thermotoga maritima (bacteria) Thermotoga maritima (bacteria) |
| Molecular weight | Theoretical: 1.3 MDa |
-Macromolecule #1: 2-dehydro-3-deoxyphosphogluconate aldolase/4-hydroxy-2-oxoglutara...
| Macromolecule | Name: 2-dehydro-3-deoxyphosphogluconate aldolase/4-hydroxy-2-oxoglutarate aldolase type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Thermotoga maritima (bacteria) Thermotoga maritima (bacteria) |
| Molecular weight | Theoretical: 23.915986 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MHHHHHHGGS GGSGGSGGSM KMEELFKKHK IVAVLRANSV EEAKKKALAV FLGGVHLIEI TFTVPDADTV IKELSFLKEM GAIIGAGTV TSVEQCRKAV ESGAEFIVSP HLDEEISQFC KEKGVFYMPG VMTPTELVKA MKLGHTILKL FPGEVVGPQF V KAMKGPFP ...String: MHHHHHHGGS GGSGGSGGSM KMEELFKKHK IVAVLRANSV EEAKKKALAV FLGGVHLIEI TFTVPDADTV IKELSFLKEM GAIIGAGTV TSVEQCRKAV ESGAEFIVSP HLDEEISQFC KEKGVFYMPG VMTPTELVKA MKLGHTILKL FPGEVVGPQF V KAMKGPFP NVKFVPTGGV NLDNVCEWFK AGVLAVGVGS ALVKGTPVEV AEKAKAFVEK IRGCTE UniProtKB: 2-dehydro-3-deoxyphosphogluconate aldolase/4-hydroxy-2-oxoglutarate aldolase |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8.1 / Details: Tris, 100 mM NaCl, 1 mM DTT |
|---|---|
| Grid | Model: Quantifoil R2/1 / Material: GOLD / Mesh: 400 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 40 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
| Details | Purified nano-cage protein at 0.1 mg/mL |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 100.0 K |
| Specialist optics | Spherical aberration corrector: Microscope was modified with a Cs corrector. |
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number grids imaged: 1 / Number real images: 1220 / Average exposure time: 69.8 sec. / Average electron dose: 44.85 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Cs: 0.05 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.2 µm / Nominal magnification: 59000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Overall B value: 69.1 / Target criteria: Correlation coefficient |
|---|---|
| Output model |  PDB-8e01: |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)