+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | 318 kDa Cas10-Csm effector complex bound to cognate target RNA | |||||||||
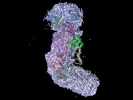
 Map data Map data | 318 kDa Cas10-Csm effector complex | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | CRISPR / Cas10 / Type III / Csm / RNA BINDING PROTEIN | |||||||||
| Biological species |  Staphylococcus epidermidis RP62A (bacteria) Staphylococcus epidermidis RP62A (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Paraan M / Stagg SM / Dunkle JA | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: PLoS One / Year: 2023 Journal: PLoS One / Year: 2023Title: The structure of a Type III-A CRISPR-Cas effector complex reveals conserved and idiosyncratic contacts to target RNA and crRNA among Type III-A systems. Authors: Mohammadreza Paraan / Mohamed Nasef / Lucy Chou-Zheng / Sarah A Khweis / Allyn J Schoeffler / Asma Hatoum-Aslan / Scott M Stagg / Jack A Dunkle /  Abstract: Type III CRISPR-Cas systems employ multiprotein effector complexes bound to small CRISPR RNAs (crRNAs) to detect foreign RNA transcripts and elicit a complex immune response that leads to the ...Type III CRISPR-Cas systems employ multiprotein effector complexes bound to small CRISPR RNAs (crRNAs) to detect foreign RNA transcripts and elicit a complex immune response that leads to the destruction of invading RNA and DNA. Type III systems are among the most widespread in nature, and emerging interest in harnessing these systems for biotechnology applications highlights the need for detailed structural analyses of representatives from diverse organisms. We performed cryo-EM reconstructions of the Type III-A Cas10-Csm effector complex from S. epidermidis bound to an intact, cognate target RNA and identified two oligomeric states, a 276 kDa complex and a 318 kDa complex. 3.1 Å density for the well-ordered 276 kDa complex allowed construction of atomic models for the Csm2, Csm3, Csm4 and Csm5 subunits within the complex along with the crRNA and target RNA. We also collected small-angle X-ray scattering data which was consistent with the 276 kDa Cas10-Csm architecture we identified. Detailed comparisons between the S. epidermidis Cas10-Csm structure and the well-resolved bacterial (S. thermophilus) and archaeal (T. onnurineus) Cas10-Csm structures reveal differences in how the complexes interact with target RNA and crRNA which are likely to have functional ramifications. These structural comparisons shed light on the unique features of Type III-A systems from diverse organisms and will assist in improving biotechnologies derived from Type III-A effector complexes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27762.map.gz emd_27762.map.gz | 230.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27762-v30.xml emd-27762-v30.xml emd-27762.xml emd-27762.xml | 19 KB 19 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_27762.png emd_27762.png | 72.8 KB | ||
| Others |  emd_27762_half_map_1.map.gz emd_27762_half_map_1.map.gz emd_27762_half_map_2.map.gz emd_27762_half_map_2.map.gz | 226.8 MB 226.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27762 http://ftp.pdbj.org/pub/emdb/structures/EMD-27762 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27762 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27762 | HTTPS FTP |
-Validation report
| Summary document |  emd_27762_validation.pdf.gz emd_27762_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_27762_full_validation.pdf.gz emd_27762_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_27762_validation.xml.gz emd_27762_validation.xml.gz | 16 KB | Display | |
| Data in CIF |  emd_27762_validation.cif.gz emd_27762_validation.cif.gz | 18.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27762 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27762 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27762 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27762 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_27762.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27762.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 318 kDa Cas10-Csm effector complex | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.846 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_27762_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
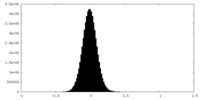
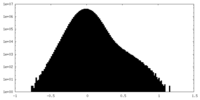
| Density Histograms |
-Half map: #1
| File | emd_27762_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
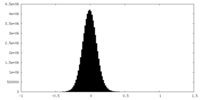
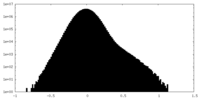
| Density Histograms |
- Sample components
Sample components
-Entire : 318 kDa Cas10-Csm effector complex bound to cognate target RNA
| Entire | Name: 318 kDa Cas10-Csm effector complex bound to cognate target RNA |
|---|---|
| Components |
|
-Supramolecule #1: 318 kDa Cas10-Csm effector complex bound to cognate target RNA
| Supramolecule | Name: 318 kDa Cas10-Csm effector complex bound to cognate target RNA type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#7 |
|---|---|
| Source (natural) | Organism:  Staphylococcus epidermidis RP62A (bacteria) Staphylococcus epidermidis RP62A (bacteria) |
| Molecular weight | Theoretical: 318 KDa |
-Macromolecule #1: Csm2
| Macromolecule | Name: Csm2 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Sequence | String: MTFAHEVVKS NVKNVKDRKG KEKQVLFNGL TTSKLRNLME QVNRLYTIAF NSNEDQLNEE FIDELEYLK IKFYYEAGRE KSVDEFLKKT LMFPIIDRVI KKESKKFFLD YCKYFEALVA Y AKYYQKED |
-Macromolecule #2: Csm3
| Macromolecule | Name: Csm3 / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Sequence | String: MYSKIKISGT IEVVTGLHIG GGGESSMIGA IDSPVVRDLQ TKLPIIPGSS IKGKMRNLLA KHFGLKMKQE SHNQDDERVL RLFGSSEKGN IQRARLQISD AFFSEKTKEH FAQNDIAYTE TKFENTINRL TAVANPRQIE RVTRGSEFDF VFIYNVDEES QVEDDFENIE ...String: MYSKIKISGT IEVVTGLHIG GGGESSMIGA IDSPVVRDLQ TKLPIIPGSS IKGKMRNLLA KHFGLKMKQE SHNQDDERVL RLFGSSEKGN IQRARLQISD AFFSEKTKEH FAQNDIAYTE TKFENTINRL TAVANPRQIE RVTRGSEFDF VFIYNVDEES QVEDDFENIE KAIHLLENDY LGGGGTRGNG RIQFKDTNIE TVVGEYDSTN LKI |
-Macromolecule #3: Csm4
| Macromolecule | Name: Csm4 / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO |
|---|---|
| Sequence | String: MTLATKVFKL SFKTPVHFGK KRLSDGEMTI TADTLFSALF IETLQLGKDT DWLLNDLIIS DTFPYENELY YLPKPLIKID SKEEDNHKAF KKLKYVPVHH YNQYLNGELS AEDATDLNDI FNIGYFSLQT KVSLIAQETD SSADSEPYSV GTFTFEPEAG LYFIAKGSEE ...String: MTLATKVFKL SFKTPVHFGK KRLSDGEMTI TADTLFSALF IETLQLGKDT DWLLNDLIIS DTFPYENELY YLPKPLIKID SKEEDNHKAF KKLKYVPVHH YNQYLNGELS AEDATDLNDI FNIGYFSLQT KVSLIAQETD SSADSEPYSV GTFTFEPEAG LYFIAKGSEE TLDHLNNIMT ALQYSGLGGK RNAGYGQFEY EIINNQQLSK LLNQNGKHSI LLSTAMAKKE EIESALKEAR YILTKRSGFV QSTNYSEMLV KKSDFYSFSS GSVFKNIFNG DIFNVGHNGK HPVYRYAKPL WLEV |
-Macromolecule #4: Csm5
| Macromolecule | Name: Csm5 / type: protein_or_peptide / ID: 4 / Enantiomer: LEVO |
|---|---|
| Sequence | String: MTIKNYEVVI KTLGPIHIGS GQVMKKQDYI YDFYNSKVYM INGNKLVKFL KRKNLLYTYQ NFLRYPPKNP RENGLKDYLD AQNVKQSEWE AFVSYSEKVN QGKKYGNTRP KPLNDLHLMV RDGQNKVYLP GSSIKGAIKT TLVSKYNNEK NKDIYSKIKV SDSKPIDESN ...String: MTIKNYEVVI KTLGPIHIGS GQVMKKQDYI YDFYNSKVYM INGNKLVKFL KRKNLLYTYQ NFLRYPPKNP RENGLKDYLD AQNVKQSEWE AFVSYSEKVN QGKKYGNTRP KPLNDLHLMV RDGQNKVYLP GSSIKGAIKT TLVSKYNNEK NKDIYSKIKV SDSKPIDESN LAIYQKIDIN KSEKSMPLYR ECIDVNTEIK FKLTIEDEIY SINEIEQSIQ DFYKNYYDKW LVGFKETKGG RRFALEGGIP DVLNQNILFL GAGTGFVSKT THYQLKNRKQ AKQDSFEILT KKFRGTYGKM KEIPSNVPVA LKGTTNQSRH TSYQQGMCKV SFQELNNEVL |
-Macromolecule #7: Cas10
| Macromolecule | Name: Cas10 / type: protein_or_peptide / ID: 7 / Enantiomer: LEVO |
|---|---|
| Sequence | String: MNKKNILMYG SLLHDIGKII YRSGDHTFSR GTHSKLGHQF LSQFSEFKDN EVLDNVAYHH YKELAKANL DNDNTAYITY IADNIASGID RRDIIEEGDE EYEKQLFNFD KYTPLYSVFN I VNSEKLKQ TNGKFKFSNE SNIEYPKTEN IQYSSGNYTT LMKDMSHDLE ...String: MNKKNILMYG SLLHDIGKII YRSGDHTFSR GTHSKLGHQF LSQFSEFKDN EVLDNVAYHH YKELAKANL DNDNTAYITY IADNIASGID RRDIIEEGDE EYEKQLFNFD KYTPLYSVFN I VNSEKLKQ TNGKFKFSNE SNIEYPKTEN IQYSSGNYTT LMKDMSHDLE HKLSIKEGTF PS LLQWTES LWQYVPSSTN KNQLIDISLY DHSRITCAIA SCIFDYLNEN NIHNYKDELF SKY ENTKSF YQKEAFLLLS MDMSGIQDFI YNISGSKALK SLRSRSFYLE LMLEVIVDQL LERL ELARA NLLYTGGGHA YLLVSNTDKV KKKITQFNNE LKKWFMSEFT TDLSLSMAFE KCSGD DLMN TSGNYRTIWR NVSSKLSDIK AHKYSAEDIL KLNHFHSYGD RECKECLRSD IDINDD GLC SICEGIINIS NDLRDKSFFV LSETGKLKMP FNKFISVIDY EEAEMLVQNN NQVRIYS KN KPYIGIGIST NLWMCDYDYA SQNQDMREKG IGSYVDREEG VKRLGVVRAD IDNLGATF I SGIPEKYNSI SRTATLSRQL SLFFKYELNH LLENYQITAI YSGGDDLFLI GAWDDIIEA SIYINDKFKE FTLDKLTLSA GVGMFSGKYP VSKMAFETGR LEEAAKTGEK NQISLWLQEK VYNWDEFKK NILEEKLLVL QQGFSQTDEH GKAFIYKMLA LLRNNEAINI ARLAYLLARS K MNEDFTSK IFNWAQNDKD KNQLITALEY YIYQIREAD |
-Macromolecule #5: crRNA
| Macromolecule | Name: crRNA / type: rna / ID: 5 |
|---|---|
| Sequence | String: ACGAGAACAC GUAUGCCGAA GUAUAUAAAU CAUCAGUACA AAG |
-Macromolecule #6: cognate target RNA
| Macromolecule | Name: cognate target RNA / type: rna / ID: 6 |
|---|---|
| Sequence | String: CUUUGUACUG AUGAUUUAUA UACUUCGGCA UACGUUCUCU AAA |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 / Component:
| ||||||
|---|---|---|---|---|---|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 78.0 K / Max: 92.0 K |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Number grids imaged: 1 / Number real images: 5000 / Average exposure time: 1.0 sec. / Average electron dose: 44.7 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Calibrated defocus max: 1.5 µm / Calibrated defocus min: 1.0 µm / Calibrated magnification: 81000 / Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Cs: 0.001 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)