[English] 日本語
 Yorodumi
Yorodumi- EMDB-27732: eEF1A(GDP)aa-tRNA stalled on the rabbit 80S ribosome by ternatin-4 -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | eEF1A(GDP)aa-tRNA stalled on the rabbit 80S ribosome by ternatin-4 | |||||||||
 Map data Map data | RELION Refine3D, pixel calibrated | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Ribosome / eEF1A / ternatin / translation | |||||||||
| Biological species |  | |||||||||
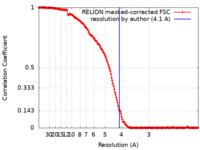
| Method | single particle reconstruction / cryo EM / Resolution: 4.1 Å | |||||||||
 Authors Authors | Brown A / Shao S / Rundlet EJ / Juette MF / Carelli JD / Taunton J / Blanchard SC | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2022 Journal: Elife / Year: 2022Title: Didemnin B and ternatin-4 differentially inhibit conformational changes in eEF1A required for aminoacyl-tRNA accommodation into mammalian ribosomes. Authors: Manuel F Juette / Jordan D Carelli / Emily J Rundlet / Alan Brown / Sichen Shao / Angelica Ferguson / Michael R Wasserman / Mikael Holm / Jack Taunton / Scott C Blanchard /   Abstract: Rapid and accurate mRNA translation requires efficient codon-dependent delivery of the correct aminoacyl-tRNA (aa-tRNA) to the ribosomal A site. In mammals, this fidelity-determining reaction is ...Rapid and accurate mRNA translation requires efficient codon-dependent delivery of the correct aminoacyl-tRNA (aa-tRNA) to the ribosomal A site. In mammals, this fidelity-determining reaction is facilitated by the GTPase elongation factor-1 alpha (eEF1A), which escorts aa-tRNA as an eEF1A(GTP)-aa-tRNA ternary complex into the ribosome. The structurally unrelated cyclic peptides didemnin B and ternatin-4 bind to the eEF1A(GTP)-aa-tRNA ternary complex and inhibit translation but have different effects on protein synthesis in vitro and in vivo. Here, we employ single-molecule fluorescence imaging and cryogenic electron microscopy to determine how these natural products inhibit translational elongation on mammalian ribosomes. By binding to a common site on eEF1A, didemnin B and ternatin-4 trap eEF1A in an intermediate state of aa-tRNA selection, preventing eEF1A release and aa-tRNA accommodation on the ribosome. We also show that didemnin B and ternatin-4 exhibit distinct effects on the dynamics of aa-tRNA selection that inform on observed disparities in their inhibition efficacies and physiological impacts. These integrated findings underscore the value of dynamics measurements in assessing the mechanism of small-molecule inhibition and highlight potential of single-molecule methods to reveal how distinct natural products differentially impact the human translation mechanism. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27732.map.gz emd_27732.map.gz | 337.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27732-v30.xml emd-27732-v30.xml emd-27732.xml emd-27732.xml | 20.1 KB 20.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_27732_fsc.xml emd_27732_fsc.xml | 16.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_27732.png emd_27732.png | 140.7 KB | ||
| Filedesc metadata |  emd-27732.cif.gz emd-27732.cif.gz | 4.7 KB | ||
| Others |  emd_27732_additional_1.map.gz emd_27732_additional_1.map.gz emd_27732_half_map_1.map.gz emd_27732_half_map_1.map.gz emd_27732_half_map_2.map.gz emd_27732_half_map_2.map.gz | 38.4 MB 338.2 MB 338 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27732 http://ftp.pdbj.org/pub/emdb/structures/EMD-27732 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27732 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27732 | HTTPS FTP |
-Validation report
| Summary document |  emd_27732_validation.pdf.gz emd_27732_validation.pdf.gz | 1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_27732_full_validation.pdf.gz emd_27732_full_validation.pdf.gz | 1 MB | Display | |
| Data in XML |  emd_27732_validation.xml.gz emd_27732_validation.xml.gz | 23.8 KB | Display | |
| Data in CIF |  emd_27732_validation.cif.gz emd_27732_validation.cif.gz | 30.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27732 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27732 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27732 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27732 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_27732.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27732.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | RELION Refine3D, pixel calibrated | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.045 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: RELION Postprocessed, pixel calibrated
| File | emd_27732_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | RELION Postprocessed, pixel calibrated | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Pixel calibrated
| File | emd_27732_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Pixel calibrated | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Pixel calibrated
| File | emd_27732_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Pixel calibrated | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : eEF1A(GDP)aa-tRNA stalled on the rabbit 80S ribosome by ternatin-4
| Entire | Name: eEF1A(GDP)aa-tRNA stalled on the rabbit 80S ribosome by ternatin-4 |
|---|---|
| Components |
|
-Supramolecule #1: eEF1A(GDP)aa-tRNA stalled on the rabbit 80S ribosome by ternatin-4
| Supramolecule | Name: eEF1A(GDP)aa-tRNA stalled on the rabbit 80S ribosome by ternatin-4 type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #2: eEF1A(GDP)-aatRNA
| Supramolecule | Name: eEF1A(GDP)-aatRNA / type: complex / ID: 2 / Parent: 1 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: Peptidyl-tRNA
| Supramolecule | Name: Peptidyl-tRNA / type: complex / ID: 3 / Parent: 1 / Details: P-site tRNA |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #4: Deacyl-tRNA
| Supramolecule | Name: Deacyl-tRNA / type: complex / ID: 4 / Parent: 1 / Details: E-site tRNA |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #5: 40S subunit
| Supramolecule | Name: 40S subunit / type: complex / ID: 5 / Parent: 1 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #6: 60S subunit
| Supramolecule | Name: 60S subunit / type: complex / ID: 6 / Parent: 1 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 Component:
Details: Included 1 uM terntin-4 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R2/2 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 5 / Pretreatment - Type: GLOW DISCHARGE | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK III | |||||||||||||||
| Details | Pull downs of ribosomes from in vitro translation reactions of a transcript encoding 3x Flag-tagged KRas in rabbit reticulocyte lysate (RRL) in the presence of 1 uM ternatin-4. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Detector mode: COUNTING / Number real images: 1422 / Average exposure time: 1.0 sec. / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 9.200000000000001 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 135000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)