+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Duplex-G-quadruplex-duplex (DGD) class_1 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | G-quadruplex / promoter / duplex / DNA | |||||||||
| Biological species | synthetic construct (others) | |||||||||
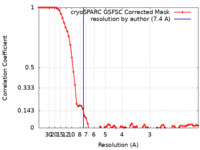
| Method | single particle reconstruction / cryo EM / Resolution: 7.4 Å | |||||||||
 Authors Authors | Monsen RC / Chua EYD / Hopkins JB / Chaires JB / Trent JO | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nucleic Acids Res / Year: 2023 Journal: Nucleic Acids Res / Year: 2023Title: Structure of a 28.5 kDa duplex-embedded G-quadruplex system resolved to 7.4 Å resolution with cryo-EM. Authors: Robert C Monsen / Eugene Y D Chua / Jesse B Hopkins / Jonathan B Chaires / John O Trent /  Abstract: Genomic regions with high guanine content can fold into non-B form DNA four-stranded structures known as G-quadruplexes (G4s). Extensive in vivo investigations have revealed that promoter G4s are ...Genomic regions with high guanine content can fold into non-B form DNA four-stranded structures known as G-quadruplexes (G4s). Extensive in vivo investigations have revealed that promoter G4s are transcriptional regulators. Little structural information exists for these G4s embedded within duplexes, their presumed genomic environment. Here, we report the 7.4 Å resolution structure and dynamics of a 28.5 kDa duplex-G4-duplex (DGD) model system using cryo-EM, molecular dynamics, and small-angle X-ray scattering (SAXS) studies. The DGD cryo-EM refined model features a 53° bend induced by a stacked duplex-G4 interaction at the 5' G-tetrad interface with a persistently unstacked 3' duplex. The surrogate complement poly dT loop preferably stacks onto the 3' G-tetrad interface resulting in occlusion of both 5' and 3' tetrad interfaces. Structural analysis shows that the DGD model is quantifiably more druggable than the monomeric G4 structure alone and represents a new structural drug target. Our results illustrate how the integration of cryo-EM, MD, and SAXS can reveal complementary detailed static and dynamic structural information on DNA G4 systems. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27726.map.gz emd_27726.map.gz | 32.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27726-v30.xml emd-27726-v30.xml emd-27726.xml emd-27726.xml | 17.7 KB 17.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_27726_fsc.xml emd_27726_fsc.xml | 6.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_27726.png emd_27726.png | 25.2 KB | ||
| Filedesc metadata |  emd-27726.cif.gz emd-27726.cif.gz | 4.9 KB | ||
| Others |  emd_27726_half_map_1.map.gz emd_27726_half_map_1.map.gz emd_27726_half_map_2.map.gz emd_27726_half_map_2.map.gz | 31.8 MB 31.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27726 http://ftp.pdbj.org/pub/emdb/structures/EMD-27726 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27726 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27726 | HTTPS FTP |
-Related structure data
| Related structure data |  8dutMC  27725  27727 M: atomic model generated by this map C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_27726.map.gz / Format: CCP4 / Size: 34.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27726.map.gz / Format: CCP4 / Size: 34.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.0252 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_27726_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
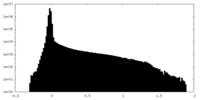
| Projections & Slices |
| ||||||||||||
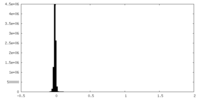
| Density Histograms |
-Half map: #2
| File | emd_27726_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
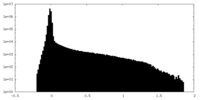
| Projections & Slices |
| ||||||||||||
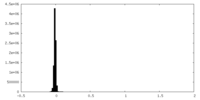
| Density Histograms |
- Sample components
Sample components
-Entire : synthetic duplex-G-quadruplex-duplex construct mimicking a promoter G4
| Entire | Name: synthetic duplex-G-quadruplex-duplex construct mimicking a promoter G4 |
|---|---|
| Components |
|
-Supramolecule #1: synthetic duplex-G-quadruplex-duplex construct mimicking a promoter G4
| Supramolecule | Name: synthetic duplex-G-quadruplex-duplex construct mimicking a promoter G4 type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
-Macromolecule #1: Duplex-G-quadruplex-duplex (46-MER)
| Macromolecule | Name: Duplex-G-quadruplex-duplex (46-MER) / type: dna / ID: 1 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 14.086031 KDa |
| Sequence | String: (DG)(DC)(DG)(DT)(DG)(DC)(DC)(DG)(DC)(DA) (DT)(DT)(DA)(DA)(DT)(DT)(DT)(DT)(DT)(DT) (DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT) (DT)(DT)(DT)(DT)(DT)(DT)(DT)(DG)(DT)(DA) (DT) (DA)(DC)(DA)(DT)(DA)(DG) |
-Macromolecule #2: Duplex-G-quadruplex-duplex (46-MER)
| Macromolecule | Name: Duplex-G-quadruplex-duplex (46-MER) / type: dna / ID: 2 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 14.407232 KDa |
| Sequence | String: (DC)(DT)(DA)(DT)(DG)(DT)(DA)(DT)(DA)(DC) (DA)(DA)(DA)(DG)(DA)(DG)(DG)(DG)(DT)(DG) (DG)(DG)(DT)(DA)(DG)(DG)(DG)(DT)(DG) (DG)(DG)(DT)(DT)(DT)(DA)(DA)(DT)(DG)(DC) (DG) (DG)(DC)(DA)(DC)(DG)(DC) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | 2D array |
- Sample preparation
Sample preparation
| Concentration | 2.5 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.2 Component:
| ||||||||||||
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: GOLD / Support film - topology: HOLEY / Support film - Film thickness: 50 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 7 sec. / Pretreatment - Atmosphere: OTHER | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 278 K / Instrument: FEI VITROBOT MARK IV | ||||||||||||
| Details | Sample was monodisperse. |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Number real images: 12175 / Average exposure time: 0.4 sec. / Average electron dose: 68.54 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.7 µm / Nominal defocus min: 0.8 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: HELIUM |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Overall B value: 557.2 / Target criteria: map correlation |
|---|---|
| Output model |  PDB-8dut: |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)