[English] 日本語
 Yorodumi
Yorodumi- EMDB-27706: Vaccine elicited Antibody MU89 bound to CH848.D949.10.17_N133D_N1... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Vaccine elicited Antibody MU89 bound to CH848.D949.10.17_N133D_N138T.DS.SOSIP.664 HIV-1 Env trimer | |||||||||
 Map data Map data | Z-Sharpened map from homogeneous C3 refinement in cryosparc (Sharpened in Phenix) | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | HIV-1 envelope / vaccine-induced antibody / broadly neutralizing antibody / VIRAL PROTEIN-IMMUNE SYSTEM complex | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /   Human immunodeficiency virus 1 / Human immunodeficiency virus 1 /   Human immunodeficiency virus / Human immunodeficiency virus /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.57 Å | |||||||||
 Authors Authors | Stalls V / Acharya P | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Cell Host Microbe / Year: 2024 Journal: Cell Host Microbe / Year: 2024Title: Mutation-guided vaccine design: A process for developing boosting immunogens for HIV broadly neutralizing antibody induction. Authors: Kevin Wiehe / Kevin O Saunders / Victoria Stalls / Derek W Cain / Sravani Venkatayogi / Joshua S Martin Beem / Madison Berry / Tyler Evangelous / Rory Henderson / Bhavna Hora / Shi-Mao Xia / ...Authors: Kevin Wiehe / Kevin O Saunders / Victoria Stalls / Derek W Cain / Sravani Venkatayogi / Joshua S Martin Beem / Madison Berry / Tyler Evangelous / Rory Henderson / Bhavna Hora / Shi-Mao Xia / Chuancang Jiang / Amanda Newman / Cindy Bowman / Xiaozhi Lu / Mary E Bryan / Joena Bal / Aja Sanzone / Haiyan Chen / Amanda Eaton / Mark A Tomai / Christopher B Fox / Ying K Tam / Christopher Barbosa / Mattia Bonsignori / Hiromi Muramatsu / S Munir Alam / David C Montefiori / Wilton B Williams / Norbert Pardi / Ming Tian / Drew Weissman / Frederick W Alt / Priyamvada Acharya / Barton F Haynes /   Abstract: A major goal of HIV-1 vaccine development is the induction of broadly neutralizing antibodies (bnAbs). Although success has been achieved in initiating bnAb B cell lineages, design of boosting ...A major goal of HIV-1 vaccine development is the induction of broadly neutralizing antibodies (bnAbs). Although success has been achieved in initiating bnAb B cell lineages, design of boosting immunogens that select for bnAb B cell receptors with improbable mutations required for bnAb affinity maturation remains difficult. Here, we demonstrate a process for designing boosting immunogens for a V3-glycan bnAb B cell lineage. The immunogens induced affinity-matured antibodies by selecting for functional improbable mutations in bnAb precursor knockin mice. Moreover, we show similar success in prime and boosting with nucleoside-modified mRNA-encoded HIV-1 envelope trimer immunogens, with improved selection by mRNA immunogens of improbable mutations required for bnAb binding to key envelope glycans. These results demonstrate the ability of both protein and mRNA prime-boost immunogens for selection of rare B cell lineage intermediates with neutralizing breadth after bnAb precursor expansion, a key proof of concept and milestone toward development of an HIV-1 vaccine. #1:  Journal: bioRxiv / Year: 2022 Journal: bioRxiv / Year: 2022Title: Mutation-Guided Vaccine Design: A Strategy for Developing Boosting Immunogens for HIV Broadly Neutralizing Antibody Induction Authors: Wiehe K / Saunders KO / Stalls V / Cain D / Venkatayogi S / Martin Beem JS / Berry M / Evangelous T / Henderson R / Hora B / Xia S / Jiang C / Newman A / Bowman C / Lu X / Bryan ME / Bal J / ...Authors: Wiehe K / Saunders KO / Stalls V / Cain D / Venkatayogi S / Martin Beem JS / Berry M / Evangelous T / Henderson R / Hora B / Xia S / Jiang C / Newman A / Bowman C / Lu X / Bryan ME / Bal J / Sanzone A / Chen H / Eaton A / Tomai MA / Fox CB / Tam Y / Barbosa C / Bonsignori M / Muramatsu H / Alam SM / Montefiori D / Williams WB / Pardi N / Tian M / Weissman D / Alt FW / Acharya P / Haynes BF | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27706.map.gz emd_27706.map.gz | 117.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27706-v30.xml emd-27706-v30.xml emd-27706.xml emd-27706.xml | 27 KB 27 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_27706.png emd_27706.png | 53.7 KB | ||
| Masks |  emd_27706_msk_1.map emd_27706_msk_1.map | 125 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-27706.cif.gz emd-27706.cif.gz | 7.7 KB | ||
| Others |  emd_27706_additional_1.map.gz emd_27706_additional_1.map.gz emd_27706_half_map_1.map.gz emd_27706_half_map_1.map.gz emd_27706_half_map_2.map.gz emd_27706_half_map_2.map.gz | 117.3 MB 115.1 MB 115.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27706 http://ftp.pdbj.org/pub/emdb/structures/EMD-27706 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27706 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27706 | HTTPS FTP |
-Validation report
| Summary document |  emd_27706_validation.pdf.gz emd_27706_validation.pdf.gz | 768.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_27706_full_validation.pdf.gz emd_27706_full_validation.pdf.gz | 768.3 KB | Display | |
| Data in XML |  emd_27706_validation.xml.gz emd_27706_validation.xml.gz | 14.1 KB | Display | |
| Data in CIF |  emd_27706_validation.cif.gz emd_27706_validation.cif.gz | 16.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27706 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27706 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27706 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27706 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_27706.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27706.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Z-Sharpened map from homogeneous C3 refinement in cryosparc (Sharpened in Phenix) | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||
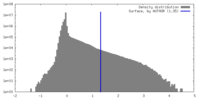
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_27706_msk_1.map emd_27706_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
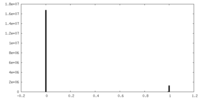
| Density Histograms |
-Additional map: Sharp map from homogeneous C3 refinement in cryosparc
| File | emd_27706_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharp map from homogeneous C3 refinement in cryosparc | ||||||||||||
| Projections & Slices |
| ||||||||||||
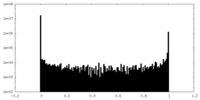
| Density Histograms |
-Half map: Half A map from homogeneous C3 refinement in cryosparc
| File | emd_27706_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half A map from homogeneous C3 refinement in cryosparc | ||||||||||||
| Projections & Slices |
| ||||||||||||
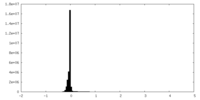
| Density Histograms |
-Half map: Half B map from homogeneous C3 refinement in cryosparc
| File | emd_27706_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half B map from homogeneous C3 refinement in cryosparc | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : V3-glycan vaccine induced antibody MU89 in complex with CH848.3.D...
| Entire | Name: V3-glycan vaccine induced antibody MU89 in complex with CH848.3.D0949.10.17chim.6R.DS.SOSIP.664_N133D_N138T HIV Env trimer |
|---|---|
| Components |
|
-Supramolecule #1: V3-glycan vaccine induced antibody MU89 in complex with CH848.3.D...
| Supramolecule | Name: V3-glycan vaccine induced antibody MU89 in complex with CH848.3.D0949.10.17chim.6R.DS.SOSIP.664_N133D_N138T HIV Env trimer type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 360 KDa |
-Macromolecule #1: CH848.3.D0949.10.17chim.6R.DS.SOSIP.664_N133D_N138T gp120
| Macromolecule | Name: CH848.3.D0949.10.17chim.6R.DS.SOSIP.664_N133D_N138T gp120 type: protein_or_peptide / ID: 1 Details: CH848.3.D0949.10.17chim.6R.DS.SOSIP.664_N133D_N138T Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 54.136465 KDa |
| Recombinant expression | Organism: Mammalia (mammals) |
| Sequence | String: MGSLQPLATL YLLGMLVASV LAAENLWVTV YYGVPVWKEA KTTLFCASDA RAYEKEVHNV WATHACVPTD PSPQELVLGN VTENFNMWK NDMVDQMHED IISLWDQSLK PCVKLTPLCV TLICSDATVK TGTVEEMKNC SFNTTTEIRD KEKKEYALFY K PDIVPLSE ...String: MGSLQPLATL YLLGMLVASV LAAENLWVTV YYGVPVWKEA KTTLFCASDA RAYEKEVHNV WATHACVPTD PSPQELVLGN VTENFNMWK NDMVDQMHED IISLWDQSLK PCVKLTPLCV TLICSDATVK TGTVEEMKNC SFNTTTEIRD KEKKEYALFY K PDIVPLSE TNNTSEYRLI NCNTSACTQA CPKVTFEPIP IHYCAPAGYA ILKCNDETFN GTGPCSNVST VQCTHGIRPV VS TQLLLNG SLAEKEIVIR SENLTNNAKI IIVHLHTPVE IVCTRPNNNT RKSVRIGPGQ TFYATGDIIG DIKQAHCNIS EEK WNDTLQ KVGIELQKHF PNKTIKYNQS AGGDMEITTH SFNCGGEFFY CNTSNLFNGT YNGTYISTNS SANSTSTITL QCRI KQIIN MWQGVGRCMY APPIAGNITC RSNITGLLLT RDGGTNSNET ETFRPAGGDM RDNWRSELYK YKVVKIEPLG VAPTR CKRR VVG |
-Macromolecule #2: Envelope Glycoprotein gp141
| Macromolecule | Name: Envelope Glycoprotein gp141 / type: protein_or_peptide / ID: 2 Details: CH848.3.D0949.10.17chim.6R.DS.SOSIP.664_N133D_N138T Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus Human immunodeficiency virus |
| Molecular weight | Theoretical: 17.146482 KDa |
| Recombinant expression | Organism: Mammalia (mammals) |
| Sequence | String: AVGIGAVFLG FLGAAGSTMG AASMTLTVQA RNLLSGIVQQ QSNLLRAPEA QQHLLKLTVW GIKQLQARVL AVERYLRDQQ LLGIWGCSG KLICCTNVPW NSSWSNRNLS EIWDNMTWLQ WDKEISNYTQ IIYGLLEESQ NQQEKNEQDL LALD |
-Macromolecule #3: MU89 Heavy Chain
| Macromolecule | Name: MU89 Heavy Chain / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 14.17273 KDa |
| Recombinant expression | Organism: Mammalia (mammals) |
| Sequence | String: QVQLVQSGAE VKKPGASVKV SCKASGYRFT DHYIHWVRQA PGQGPEWMGW INTSSGRSNF AQKFQGRVTM TRDTSISTAY MELNRLKSD DTAVYYCTTG SWISLYYDSS GYPNFDYWGQ GTLVTVT |
-Macromolecule #4: MU89 Light Chain
| Macromolecule | Name: MU89 Light Chain / type: protein_or_peptide / ID: 4 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 10.989062 KDa |
| Recombinant expression | Organism: Mammalia (mammals) |
| Sequence | String: TQPASVSGSP GQPITISCTG TSSDVGNYDL VSWYQQHPGN APKYMIYEVT KRPAGISNRF SGSKSGNTAS LTISGLQAED AADYYCCSY AGSSTVIFGG GTKVTVL |
-Macromolecule #11: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 11 / Number of copies: 12 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.5 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number real images: 1787 / Average electron dose: 58.9 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.7000000000000001 µm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)