+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the PEAK3 pseudokinase homodimer | ||||||||||||
 Map data Map data | Reconstruction of the PEAK3 pseudokinase homodimer filtered to 4.9A and sharpened with -105 bFactor. | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | complex / pseudokinase / kinase / adapter / SIGNALING PROTEIN | ||||||||||||
| Function / homology | : / regulation of cell motility / regulation of actin cytoskeleton organization / actin cytoskeleton / regulation of cell shape / protein kinase activity / focal adhesion / Protein PEAK3 Function and homology information Function and homology information | ||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.9 Å | ||||||||||||
 Authors Authors | Torosyan H / Paul M / Jura N / Verba KA | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Structural insights into regulation of the PEAK3 pseudokinase scaffold by 14-3-3. Authors: Torosyan H / Paul MD / Forget A / Lo M / Diwanji D / Pawlowski K / Krogan NJ / Jura N / Verba KA | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27684.map.gz emd_27684.map.gz | 7.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27684-v30.xml emd-27684-v30.xml emd-27684.xml emd-27684.xml | 18 KB 18 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_27684_fsc.xml emd_27684_fsc.xml | 11.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_27684.png emd_27684.png | 60.1 KB | ||
| Masks |  emd_27684_msk_1.map emd_27684_msk_1.map | 125 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-27684.cif.gz emd-27684.cif.gz | 6.2 KB | ||
| Others |  emd_27684_half_map_1.map.gz emd_27684_half_map_1.map.gz emd_27684_half_map_2.map.gz emd_27684_half_map_2.map.gz | 98.3 MB 98.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27684 http://ftp.pdbj.org/pub/emdb/structures/EMD-27684 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27684 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27684 | HTTPS FTP |
-Validation report
| Summary document |  emd_27684_validation.pdf.gz emd_27684_validation.pdf.gz | 826.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_27684_full_validation.pdf.gz emd_27684_full_validation.pdf.gz | 826.1 KB | Display | |
| Data in XML |  emd_27684_validation.xml.gz emd_27684_validation.xml.gz | 18.5 KB | Display | |
| Data in CIF |  emd_27684_validation.cif.gz emd_27684_validation.cif.gz | 24.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27684 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27684 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27684 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27684 | HTTPS FTP |
-Related structure data
| Related structure data | 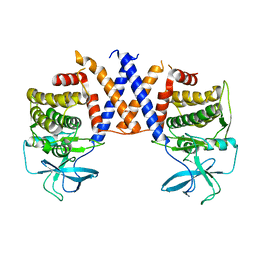 8ds6MC  8dp5C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_27684.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27684.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction of the PEAK3 pseudokinase homodimer filtered to 4.9A and sharpened with -105 bFactor. | ||||||||||||||||||||
| Voxel size | X=Y=Z: 0.835 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : PEAK3 pseudokinase homodimer
| Entire | Name: PEAK3 pseudokinase homodimer |
|---|---|
| Components |
|
-Supramolecule #1: PEAK3 pseudokinase homodimer
| Supramolecule | Name: PEAK3 pseudokinase homodimer / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 104.58 KDa |
-Macromolecule #1: Protein PEAK3
| Macromolecule | Name: Protein PEAK3 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 52.357031 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MSSPEPPTEP PEPDNPTWST QPTYSNLGQI RAHLLPSKAC RLRTPGSLST NPEPLPPPLP KKILTRTQSL PTRRTLHPSS IQVQPPRRP FLGSHSVDKS QAAVGPACLP AELTFGPADA PLGLSLRDLH SPEAVHTALA ARQLQGLRTI YARLRARLMG G HPGPCHPG ...String: MSSPEPPTEP PEPDNPTWST QPTYSNLGQI RAHLLPSKAC RLRTPGSLST NPEPLPPPLP KKILTRTQSL PTRRTLHPSS IQVQPPRRP FLGSHSVDKS QAAVGPACLP AELTFGPADA PLGLSLRDLH SPEAVHTALA ARQLQGLRTI YARLRARLMG G HPGPCHPG HSFRLLDSSP CAESGDALYY RVVRAHEDAW HILVAKVPKP GADVPHPWGL ELQASLSPHF NLQGLCGLVP EG TLPGAPW RGAVALAAEV PERTVAQWLA EACTQPPEEF VWAVALLLLQ LSAALKFLEA WGAALVELRP ENLLLVAPRG CAT TGPPRL LLTDFGRVCL QPPGPPGSPG PHAPQLGSLL RALLSLAAPS TTPLAAGLEL LAAQLTRLRP SASRTRGALQ ALLW GPGPE LRGRGAPLGP WLRALGPWLR VRRGLLVLRL AERAAGGEAP SLEDWLCCEY LAEATESSMG QALALLWDLE GGGGA DYKD DDDKGPV UniProtKB: Protein PEAK3 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.1 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
Details: A final concentration of 0.1% of Octyl-beta-Glucoside (C14H28O6) was added to the sample before freezing. | |||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 278.15 K / Instrument: FEI VITROBOT MARK IV / Details: blot time = 7s blot force = 4. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Average electron dose: 69.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 4.9 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION (ver. 3.1.0) / Number images used: 32734 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: cryoSPARC (ver. 2.15) |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller




