+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Quorum-sensing receptor RhlR bound to PqsE | |||||||||||||||
 Map data Map data | Density modified full map | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | transcriptional regulators / quorum sensing / DNA binding / TRANSCRIPTION | |||||||||||||||
| Function / homology |  Function and homology information Function and homology information2-aminobenzoylacetyl-CoA thioesterase / secondary metabolite biosynthetic process / hydrolase activity / regulation of DNA-templated transcription / DNA binding / metal ion binding Similarity search - Function | |||||||||||||||
| Biological species |  | |||||||||||||||
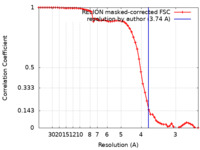
| Method | single particle reconstruction / cryo EM / Resolution: 3.74 Å | |||||||||||||||
 Authors Authors | Paczkowski JE / Fromme JC / Feathers JR | |||||||||||||||
| Funding support |  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Structure / Year: 2022 Journal: Structure / Year: 2022Title: Structure of the RhlR-PqsE complex from Pseudomonas aeruginosa reveals mechanistic insights into quorum-sensing gene regulation. Authors: J Ryan Feathers / Erica K Richael / Kayla A Simanek / J Christopher Fromme / Jon E Paczkowski /  Abstract: Pseudomonas aeruginosa is an opportunistic pathogen that is responsible for thousands of deaths every year in the United States. P. aeruginosa virulence factor production is mediated by quorum ...Pseudomonas aeruginosa is an opportunistic pathogen that is responsible for thousands of deaths every year in the United States. P. aeruginosa virulence factor production is mediated by quorum sensing, a mechanism of bacterial cell-cell communication that relies on the production and detection of signal molecules called autoinducers. In P. aeruginosa, the transcription factor receptor RhlR is activated by a RhlI-synthesized autoinducer. We recently showed that RhlR-dependent transcription is enhanced by a physical interaction with the enzyme PqsE via increased affinity of RhlR for promoter DNA. However, the molecular basis for complex formation and how complex formation enhanced RhlR transcriptional activity remained unclear. Here, we report the structure of ligand-bound RhlR in complex with PqsE. Additionally, we determined the structure of the complex bound with DNA, revealing the mechanism by which RhlR-mediated transcription is enhanced by PqsE, thereby establishing the molecular basis for RhlR-dependent virulence factor production in P. aeruginosa. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27645.map.gz emd_27645.map.gz | 1.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27645-v30.xml emd-27645-v30.xml emd-27645.xml emd-27645.xml | 29 KB 29 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_27645_fsc.xml emd_27645_fsc.xml | 6 KB | Display |  FSC data file FSC data file |
| Images |  emd_27645.png emd_27645.png | 43.5 KB | ||
| Filedesc metadata |  emd-27645.cif.gz emd-27645.cif.gz | 6.7 KB | ||
| Others |  emd_27645_additional_1.map.gz emd_27645_additional_1.map.gz emd_27645_additional_2.map.gz emd_27645_additional_2.map.gz emd_27645_additional_3.map.gz emd_27645_additional_3.map.gz emd_27645_additional_4.map.gz emd_27645_additional_4.map.gz emd_27645_half_map_1.map.gz emd_27645_half_map_1.map.gz emd_27645_half_map_2.map.gz emd_27645_half_map_2.map.gz | 1.6 MB 13.8 MB 1.6 MB 1.4 MB 13.8 MB 13.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27645 http://ftp.pdbj.org/pub/emdb/structures/EMD-27645 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27645 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27645 | HTTPS FTP |
-Validation report
| Summary document |  emd_27645_validation.pdf.gz emd_27645_validation.pdf.gz | 747.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_27645_full_validation.pdf.gz emd_27645_full_validation.pdf.gz | 747.2 KB | Display | |
| Data in XML |  emd_27645_validation.xml.gz emd_27645_validation.xml.gz | 11.6 KB | Display | |
| Data in CIF |  emd_27645_validation.cif.gz emd_27645_validation.cif.gz | 15.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27645 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27645 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27645 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27645 | HTTPS FTP |
-Related structure data
| Related structure data |  8dq0MC  8dq1C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_27645.map.gz / Format: CCP4 / Size: 18.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27645.map.gz / Format: CCP4 / Size: 18.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Density modified full map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.29 Å | ||||||||||||||||||||||||||||||||||||
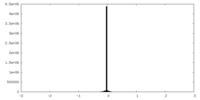
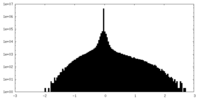
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Density modified half map 2
| File | emd_27645_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Density modified half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
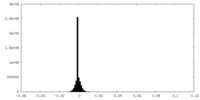
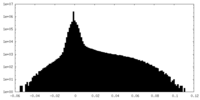
| Density Histograms |
-Additional map: Refinement full map
| File | emd_27645_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Refinement full map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Density modified half map 1
| File | emd_27645_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Density modified half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Sharpened refinement map
| File | emd_27645_additional_4.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened refinement map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Refinement unfiltered half map 2
| File | emd_27645_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Refinement unfiltered half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Refinement unfiltered half map 1
| File | emd_27645_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Refinement unfiltered half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Tetrameric complex of RhlR:mBTL bound to PqsE
| Entire | Name: Tetrameric complex of RhlR:mBTL bound to PqsE |
|---|---|
| Components |
|
-Supramolecule #1: Tetrameric complex of RhlR:mBTL bound to PqsE
| Supramolecule | Name: Tetrameric complex of RhlR:mBTL bound to PqsE / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 123 KDa |
-Macromolecule #1: RhlR protein
| Macromolecule | Name: RhlR protein / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 27.611596 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MRNDGGFLLW WDGLRSEMQP IHDSQGVFAV LEKEVRRLGF DYYAYGVRHT IPFTRPKTEV HGTYPKAWLE RYQMQNYGAV DPAILNGLR SSEMVVWSDS LFDQSRMLWN EARDWGLCVG ATLPIRAPNN LLSVLSVARD QQNISSFERE EIRLRLRCMI E LLTQKLTD ...String: MRNDGGFLLW WDGLRSEMQP IHDSQGVFAV LEKEVRRLGF DYYAYGVRHT IPFTRPKTEV HGTYPKAWLE RYQMQNYGAV DPAILNGLR SSEMVVWSDS LFDQSRMLWN EARDWGLCVG ATLPIRAPNN LLSVLSVARD QQNISSFERE EIRLRLRCMI E LLTQKLTD LEHPMLMSNP VCLSHREREI LQWTADGKSS GEIAIILSIS ESTVNFHHKN IQKKFDAPNK TLAAAYAAAL GL I UniProtKB: Regulatory protein RhlR |
-Macromolecule #2: 2-aminobenzoylacetyl-CoA thioesterase
| Macromolecule | Name: 2-aminobenzoylacetyl-CoA thioesterase / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO / EC number: 2-aminobenzoylacetyl-CoA thioesterase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 34.350297 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MLRLSAPGQL DDDLCLLGDV QVPVFLLRLG EASWALVEGG ISRDAELVWA DLCRWVADPS QVHYWLITHK HYDHCGLLPY LCPRLPNVQ VLASERTCQA WKSESAVRVV ERLNRQLLRA EQRLPEACAW DALPVRAVAD GEWLELGPRH RLQVIEAHGH S DDHVVFYD ...String: MLRLSAPGQL DDDLCLLGDV QVPVFLLRLG EASWALVEGG ISRDAELVWA DLCRWVADPS QVHYWLITHK HYDHCGLLPY LCPRLPNVQ VLASERTCQA WKSESAVRVV ERLNRQLLRA EQRLPEACAW DALPVRAVAD GEWLELGPRH RLQVIEAHGH S DDHVVFYD VRRRRLFCGD ALGEFDEAEG VWRPLVFDDM EAYLESLERL QRLPTLLQLI PGHGGLLRGR LAADGAESAY TE CLRLCRR LLWRQSMGES LDELSEELHR AWGGQSVDFL PGELHLGSMR RMLEILSRQA LPLD UniProtKB: 2-aminobenzoylacetyl-CoA thioesterase |
-Macromolecule #3: 4-(3-bromophenoxy)-N-[(3S)-2-oxothiolan-3-yl]butanamide
| Macromolecule | Name: 4-(3-bromophenoxy)-N-[(3S)-2-oxothiolan-3-yl]butanamide type: ligand / ID: 3 / Number of copies: 2 / Formula: K5G |
|---|---|
| Molecular weight | Theoretical: 358.251 Da |
| Chemical component information | 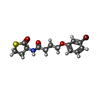 ChemComp-K5G: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5.0 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)