[English] 日本語
 Yorodumi
Yorodumi- EMDB-27625: Helical reconstruction of A92E HIV capsid in presence of FG mutan... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Helical reconstruction of A92E HIV capsid in presence of FG mutant CPSF6 construct (filtered by local resolution) | |||||||||
 Map data Map data | Helical reconstruction of A92E HIV capsid in presence of FG mutant CPSF6 construct (filtered by local resolution) | |||||||||
 Sample Sample |
| |||||||||
| Biological species |   Human immunodeficiency virus 1 Human immunodeficiency virus 1 | |||||||||
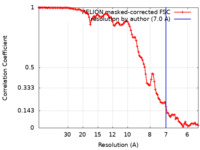
| Method | helical reconstruction / cryo EM / Resolution: 7.0 Å | |||||||||
 Authors Authors | Iqbal N / Asturias F / Kvaratskhelia M | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Prion-like low complexity regions enable avid virus-host interactions during HIV-1 infection. Authors: Guochao Wei / Naseer Iqbal / Valentine V Courouble / Ashwanth C Francis / Parmit K Singh / Arpa Hudait / Arun S Annamalai / Stephanie Bester / Szu-Wei Huang / Nikoloz Shkriabai / Lorenzo ...Authors: Guochao Wei / Naseer Iqbal / Valentine V Courouble / Ashwanth C Francis / Parmit K Singh / Arpa Hudait / Arun S Annamalai / Stephanie Bester / Szu-Wei Huang / Nikoloz Shkriabai / Lorenzo Briganti / Reed Haney / Vineet N KewalRamani / Gregory A Voth / Alan N Engelman / Gregory B Melikyan / Patrick R Griffin / Francisco Asturias / Mamuka Kvaratskhelia /  Abstract: Cellular proteins CPSF6, NUP153 and SEC24C play crucial roles in HIV-1 infection. While weak interactions of short phenylalanine-glycine (FG) containing peptides with isolated capsid hexamers have ...Cellular proteins CPSF6, NUP153 and SEC24C play crucial roles in HIV-1 infection. While weak interactions of short phenylalanine-glycine (FG) containing peptides with isolated capsid hexamers have been characterized, how these cellular factors functionally engage with biologically relevant mature HIV-1 capsid lattices is unknown. Here we show that prion-like low complexity regions (LCRs) enable avid CPSF6, NUP153 and SEC24C binding to capsid lattices. Structural studies revealed that multivalent CPSF6 assembly is mediated by LCR-LCR interactions, which are templated by binding of CPSF6 FG peptides to a subset of hydrophobic capsid pockets positioned along adjoining hexamers. In infected cells, avid CPSF6 LCR-mediated binding to HIV-1 cores is essential for functional virus-host interactions. The investigational drug lenacapavir accesses unoccupied hydrophobic pockets in the complex to potently impair HIV-1 inside the nucleus without displacing the tightly bound cellular cofactor from virus cores. These results establish previously undescribed mechanisms of virus-host interactions and antiviral action. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27625.map.gz emd_27625.map.gz | 24.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27625-v30.xml emd-27625-v30.xml emd-27625.xml emd-27625.xml | 15.5 KB 15.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_27625_fsc.xml emd_27625_fsc.xml | 10.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_27625.png emd_27625.png | 64.9 KB | ||
| Others |  emd_27625_half_map_1.map.gz emd_27625_half_map_1.map.gz emd_27625_half_map_2.map.gz emd_27625_half_map_2.map.gz | 80.8 MB 80.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27625 http://ftp.pdbj.org/pub/emdb/structures/EMD-27625 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27625 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27625 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_27625.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27625.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Helical reconstruction of A92E HIV capsid in presence of FG mutant CPSF6 construct (filtered by local resolution) | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.8 Å | ||||||||||||||||||||||||||||||||||||
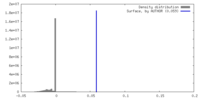
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half Map 1
| File | emd_27625_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 1 | ||||||||||||
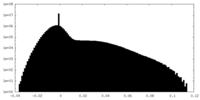
| Projections & Slices |
| ||||||||||||
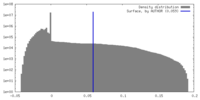
| Density Histograms |
-Half map: Half Map 2
| File | emd_27625_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
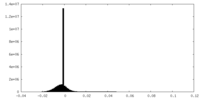
| Density Histograms |
- Sample components
Sample components
-Entire : HIV Capsid Protein Hexamer
| Entire | Name: HIV Capsid Protein Hexamer |
|---|---|
| Components |
|
-Supramolecule #1: HIV Capsid Protein Hexamer
| Supramolecule | Name: HIV Capsid Protein Hexamer / type: complex / ID: 1 / Chimera: Yes / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
-Macromolecule #1: HIV Capsid
| Macromolecule | Name: HIV Capsid / type: protein_or_peptide / ID: 1 / Enantiomer: DEXTRO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Recombinant expression | Organism:  |
| Sequence | String: PIVQNLQGQM VHQCISPRTL NAWVKVVEEK AFSPEVIPMF SALSCGATPQ DLNTMLNTVG GHQAAMQMLK ETINEEAAEW DRLHPVHAGP IAPGQMREPR GSDIAGTTST LQEQIGWMTH NPPIPVGEIY KRWIILGLNK IVRMYSPTSI LDIRQGPKEP FRDYVDRFYK ...String: PIVQNLQGQM VHQCISPRTL NAWVKVVEEK AFSPEVIPMF SALSCGATPQ DLNTMLNTVG GHQAAMQMLK ETINEEAAEW DRLHPVHAGP IAPGQMREPR GSDIAGTTST LQEQIGWMTH NPPIPVGEIY KRWIILGLNK IVRMYSPTSI LDIRQGPKEP FRDYVDRFYK TLRAEQASQE VKNAATETLL VQNANPDCKT ILKALGPGAT LEEMMTACQG VGGPGHKARV L |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | 3D array |
- Sample preparation
Sample preparation
| Concentration | 3 mg/mL |
|---|---|
| Buffer | pH: 8 / Details: 1M NaCl, 50mM Tris pH 8, 0.2mM IP6 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Number grids imaged: 2 / Number real images: 2157 / Average electron dose: 45.65 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 28000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 28000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)