[English] 日本語
 Yorodumi
Yorodumi- EMDB-27622: Vaccine Elicited Antibody DH1285 Fab bound to CH505TFchim.6R.SOSI... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Vaccine Elicited Antibody DH1285 Fab bound to CH505TFchim.6R.SOSIP.664.v4.1 Trimer, 2-Fab-bound configuration | |||||||||
 Map data Map data | Sharp map from homogeneous C1 refinement in cryosparc | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Antibody / IgG / HIV-1 / CD4 binding site / gp120 / Env / Fab / DH1285 / VIRAL PROTEIN | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /   Human immunodeficiency virus 1 / Human immunodeficiency virus 1 /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 5.5 Å | |||||||||
 Authors Authors | Stalls V / Achyara P | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: To be published Journal: To be publishedTitle: Rhesus macaque vaccine elicited antibody DH1285 bound to HIV-1 Env trimer Authors: Stalls V / Acharya P | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27622.map.gz emd_27622.map.gz | 117.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27622-v30.xml emd-27622-v30.xml emd-27622.xml emd-27622.xml | 16.7 KB 16.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_27622.png emd_27622.png | 44.6 KB | ||
| Others |  emd_27622_half_map_1.map.gz emd_27622_half_map_1.map.gz emd_27622_half_map_2.map.gz emd_27622_half_map_2.map.gz | 115.6 MB 115.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27622 http://ftp.pdbj.org/pub/emdb/structures/EMD-27622 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27622 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27622 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_27622.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27622.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharp map from homogeneous C1 refinement in cryosparc | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||
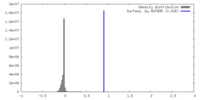
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half B map from homogeneous C1 refinement in cryosparc
| File | emd_27622_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half B map from homogeneous C1 refinement in cryosparc | ||||||||||||
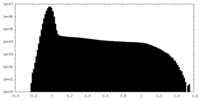
| Projections & Slices |
| ||||||||||||
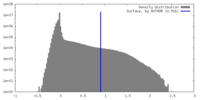
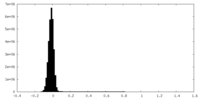
| Density Histograms |
-Half map: Half A map from homogeneous C1 refinement in cryosparc
| File | emd_27622_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half A map from homogeneous C1 refinement in cryosparc | ||||||||||||
| Projections & Slices |
| ||||||||||||
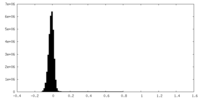
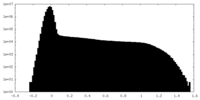
| Density Histograms |
- Sample components
Sample components
-Entire : Vaccine Elicited Antibody DH1285 Fab bound to CH505TFchim.6R.SOSI...
| Entire | Name: Vaccine Elicited Antibody DH1285 Fab bound to CH505TFchim.6R.SOSIP.664.v4.1. Trimer, 2-Fab-bound configuration |
|---|---|
| Components |
|
-Supramolecule #1: Vaccine Elicited Antibody DH1285 Fab bound to CH505TFchim.6R.SOSI...
| Supramolecule | Name: Vaccine Elicited Antibody DH1285 Fab bound to CH505TFchim.6R.SOSIP.664.v4.1. Trimer, 2-Fab-bound configuration type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 308 KDa |
-Macromolecule #1: CH505TFchim.6R.SOSIP.664.V4.1 Env Trimer
| Macromolecule | Name: CH505TFchim.6R.SOSIP.664.V4.1 Env Trimer / type: other / ID: 1 / Classification: other |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Sequence | String: MPMGSLQPLA TLYLLGMLVA SVLAAENLWV TVYYGVPVWK EAKTTLFCAS DAKAYEKKVH NVWATHACVP TDPNPQEMVL KNVTENFNMW KNDMVDQMHE DVISLWDQSL KPCVKLTPLC VTLNCTNATA SNSSIIEGMK NCSFNITTEL RDKREKKNAL FYKLDIVQLD ...String: MPMGSLQPLA TLYLLGMLVA SVLAAENLWV TVYYGVPVWK EAKTTLFCAS DAKAYEKKVH NVWATHACVP TDPNPQEMVL KNVTENFNMW KNDMVDQMHE DVISLWDQSL KPCVKLTPLC VTLNCTNATA SNSSIIEGMK NCSFNITTEL RDKREKKNAL FYKLDIVQLD GNSSQYRLIN CNTSVITQAC PKVSFDPIPI HYCAPAGYAI LKCNNKTFTG TGPCNNVSTV QCTHGIKPVV STQLLLNGSL AEGEIIIRSE NITNNVKTII VHLNESVKIE CTRPNNKTRT SIRIGPGQWF YATGQVIGDI REAYCNINES KWNETLQRVS KKLKEYFPHK NITFQPSSGG DLEITTHSFN CGGEFFYCNT SSLFNRTYMA NSTDMANSTE TNSTRTITIH CRIKQIINMW QEVGRAMYAP PIAGNITCIS NITGLLLTRD GGKNNTETFR PGGGNMKDNW RSELYKYKVV KIEPLGVAPT RCKRRVVGRR RRRRAVGIGA VFLGFLGAAG STMGAASMTL TVQARNLLSG IVQQQSNLLR APEAQQHLLK LTVWGIKQLQ ARVLAVERYL RDQQLLGIWG CSGKLICCTN VPWNSSWSNR NLSEIWDNMT WLQWDKEISN YTQIIYGLLE ESQNQQEKNE QDLLALD |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #2: DH1285 Fab Heavy Chain
| Macromolecule | Name: DH1285 Fab Heavy Chain / type: other / ID: 2 / Classification: other |
|---|---|
| Source (natural) | Organism:  |
| Sequence | String: MGWSCIILFL VATATGVHAQ VHLEQSGAEV KEPGSSVRLS CEASGYTFTD YYIHWVRQSP RQGLEWMGWI NPYYGNTHYA EKFQGRVAMT RDRSTTTAYM DLSSLTSEDT AVYYCARDEG GSGSYSYFDS WGQGVLVTVS SASTKGPSVF PLAPSSRSTS ESTAALGCLV ...String: MGWSCIILFL VATATGVHAQ VHLEQSGAEV KEPGSSVRLS CEASGYTFTD YYIHWVRQSP RQGLEWMGWI NPYYGNTHYA EKFQGRVAMT RDRSTTTAYM DLSSLTSEDT AVYYCARDEG GSGSYSYFDS WGQGVLVTVS SASTKGPSVF PLAPSSRSTS ESTAALGCLV KDYFPEPVTV SWNSGSLTSG VHTFPAVLQS SGLYSLSSVV TVPSSSLGTQ TYVCNVNHKP SNTKVDKRVE IKT |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #3: DH1285 Fab Light Chain
| Macromolecule | Name: DH1285 Fab Light Chain / type: other / ID: 3 / Classification: other |
|---|---|
| Source (natural) | Organism:  |
| Sequence | String: MGWSCIILFL VATATGVHAD IQMTQSPSSL SASVGDTVTI TCRASQDINN HLSWYQQKPG RAPKALIYSA SSLETGVPSR FSGSGSGTDY TLTISSLQPE DFATYYCQHY STSPYTFGRG TKVDIKRAVA APSVFIFPPS EDQVKSGTVS VVCLLNNFYP REASVKWKVD ...String: MGWSCIILFL VATATGVHAD IQMTQSPSSL SASVGDTVTI TCRASQDINN HLSWYQQKPG RAPKALIYSA SSLETGVPSR FSGSGSGTDY TLTISSLQPE DFATYYCQHY STSPYTFGRG TKVDIKRAVA APSVFIFPPS EDQVKSGTVS VVCLLNNFYP REASVKWKVD GVLKTGNSQE SVTEQDSKDN TYSLSSTLTL SSTDYQSHNV YACEVTHQGL SSPVTKSFNR GEC |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 59.1 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.4 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL |
|---|
 Movie
Movie Controller
Controller



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)