[English] 日本語
 Yorodumi
Yorodumi- EMDB-27573: Acidipropionibacterium acidipropionici encapsulin in an open stat... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Acidipropionibacterium acidipropionici encapsulin in an open state at pH 7.5 | |||||||||
 Map data Map data | Acidipropionibacterium acidipropionici encapsulin in an open state at pH 7.5 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Encapsulin / VIRUS LIKE PARTICLE | |||||||||
| Function / homology | Type 1 encapsulin shell protein / Encapsulating protein for peroxidase / : / encapsulin nanocompartment / Type 1 encapsulin shell protein Function and homology information Function and homology information | |||||||||
| Biological species |  Acidipropionibacterium acidipropionici ATCC 4875 (bacteria) Acidipropionibacterium acidipropionici ATCC 4875 (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.32 Å | |||||||||
 Authors Authors | Jones JA / Andreas MP / Giessen TW | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Biomacromolecules / Year: 2023 Journal: Biomacromolecules / Year: 2023Title: Exploring the Extreme Acid Tolerance of a Dynamic Protein Nanocage. Authors: Jesse A Jones / Michael P Andreas / Tobias W Giessen /  Abstract: Encapsulins are microbial protein nanocages capable of efficient self-assembly and cargo enzyme encapsulation. Due to their favorable properties, including high thermostability, protease resistance, ...Encapsulins are microbial protein nanocages capable of efficient self-assembly and cargo enzyme encapsulation. Due to their favorable properties, including high thermostability, protease resistance, and robust heterologous expression, encapsulins have become popular bioengineering tools for applications in medicine, catalysis, and nanotechnology. Resistance against physicochemical extremes like high temperature and low pH is a highly desirable feature for many biotechnological applications. However, no systematic search for acid-stable encapsulins has been carried out, while the influence of pH on encapsulin shells has so far not been thoroughly explored. Here, we report on a newly identified encapsulin nanocage from the acid-tolerant bacterium . Using transmission electron microscopy, dynamic light scattering, and proteolytic assays, we demonstrate its extreme acid tolerance and resilience against proteases. We structurally characterize the novel nanocage using cryo-electron microscopy, revealing a dynamic five-fold pore that displays distinct "closed" and "open" states at neutral pH but only a singular "closed" state under strongly acidic conditions. Further, the "open" state exhibits the largest pore in an encapsulin shell reported to date. Non-native protein encapsulation capabilities are demonstrated, and the influence of external pH on internalized cargo is explored. Our results expand the biotechnological application range of encapsulin nanocages toward potential uses under strongly acidic conditions and highlight pH-responsive encapsulin pore dynamics. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27573.map.gz emd_27573.map.gz | 204.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27573-v30.xml emd-27573-v30.xml emd-27573.xml emd-27573.xml | 17.8 KB 17.8 KB | Display Display |  EMDB header EMDB header |
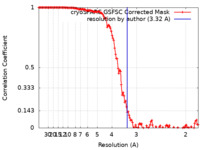
| FSC (resolution estimation) |  emd_27573_fsc.xml emd_27573_fsc.xml | 12.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_27573.png emd_27573.png | 154.3 KB | ||
| Filedesc metadata |  emd-27573.cif.gz emd-27573.cif.gz | 5.9 KB | ||
| Others |  emd_27573_half_map_1.map.gz emd_27573_half_map_1.map.gz emd_27573_half_map_2.map.gz emd_27573_half_map_2.map.gz | 200.4 MB 200.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27573 http://ftp.pdbj.org/pub/emdb/structures/EMD-27573 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27573 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27573 | HTTPS FTP |
-Related structure data
| Related structure data |  8dnlMC  8dn9C  8dnaC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_27573.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27573.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Acidipropionibacterium acidipropionici encapsulin in an open state at pH 7.5 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.91 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Acidipropionibacterium acidipropionici encapsulin in an open state at...
| File | emd_27573_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Acidipropionibacterium acidipropionici encapsulin in an open state at pH 7.5 | ||||||||||||
| Projections & Slices |
| ||||||||||||
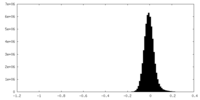
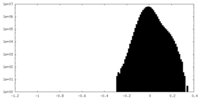
| Density Histograms |
-Half map: Acidipropionibacterium acidipropionici encapsulin in an open state at...
| File | emd_27573_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Acidipropionibacterium acidipropionici encapsulin in an open state at pH 7.5 | ||||||||||||
| Projections & Slices |
| ||||||||||||
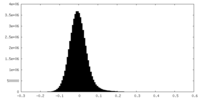
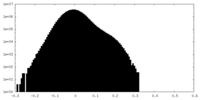
| Density Histograms |
- Sample components
Sample components
-Entire : Acidipropionibacterium acidipropionici encapsulin in an open stat...
| Entire | Name: Acidipropionibacterium acidipropionici encapsulin in an open state at pH 7.5 |
|---|---|
| Components |
|
-Supramolecule #1: Acidipropionibacterium acidipropionici encapsulin in an open stat...
| Supramolecule | Name: Acidipropionibacterium acidipropionici encapsulin in an open state at pH 7.5 type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Acidipropionibacterium acidipropionici ATCC 4875 (bacteria) Acidipropionibacterium acidipropionici ATCC 4875 (bacteria) |
| Molecular weight | Theoretical: 1.71 MDa |
-Macromolecule #1: 29 kDa antigen cfp29
| Macromolecule | Name: 29 kDa antigen cfp29 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Acidipropionibacterium acidipropionici ATCC 4875 (bacteria) Acidipropionibacterium acidipropionici ATCC 4875 (bacteria)Strain: ATCC 4875 / DSM 20272 / JCM 6432 / NBRC 12425 / NCIMB 8070 / 4 |
| Molecular weight | Theoretical: 28.522725 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MNNLHRELAP ISEAAWKQID DEARDTFSLR AAGRRVVDVP EPAGPTLGSV SLGHLETGSQ TDGVQTSVYR VQPLVQVRVP FTVSRADID DVERGAVDLT WDPVDDAVAK LVDTEDTAIL HGWEEAGITG LSEASVHQPV QMPAELEQID DAVSGACNVL R LADVEGPY ...String: MNNLHRELAP ISEAAWKQID DEARDTFSLR AAGRRVVDVP EPAGPTLGSV SLGHLETGSQ TDGVQTSVYR VQPLVQVRVP FTVSRADID DVERGAVDLT WDPVDDAVAK LVDTEDTAIL HGWEEAGITG LSEASVHQPV QMPAELEQID DAVSGACNVL R LADVEGPY DLVLPQQLYT QVSETTDHGV PVVDHLTQLL SGGEVLWAPA ARCALVVSRR GGDSCLFLGR DVSIGYLSHD AQ TVTLYLE ESFTFRVHQP DAAVALV UniProtKB: Type 1 encapsulin shell protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3.0 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR Details: The grid was glow discharged for 60 seconds at 5 mA. | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 295 K / Instrument: FEI VITROBOT MARK IV Details: Additional settings were : Blot force- 20, blot time - 4 seconds, drain time - 0 seconds, wait time - 0 seconds. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Number grids imaged: 1 / Number real images: 975 / Average exposure time: 4.0 sec. / Average electron dose: 41.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 45000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Details | A starting model was created using RosettaFold. The starting model was placed in the map manually using UCSF Chimera. The model was then further refined using Coot and Phenix Real Space Refine. |
|---|---|
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Overall B value: 77.81 / Target criteria: Correlation coefficient |
| Output model |  PDB-8dnl: |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)