[English] 日本語
 Yorodumi
Yorodumi- EMDB-27556: CryoEM structure of the A. aeolicus WzmWzt transporter bound to 3... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of the A. aeolicus WzmWzt transporter bound to 3-O-methyl-D-mannose | |||||||||
 Map data Map data | sharpened | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | O antigen ABC transporter / 3-O-methyl-D-mannose / TRANSLOCASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationpolysaccharide transport / lipopolysaccharide transport / ABC-type transporter activity / ATP hydrolysis activity / ATP binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |   Aquifex aeolicus (bacteria) Aquifex aeolicus (bacteria) | |||||||||
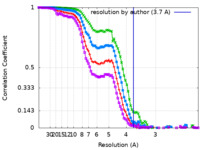
| Method | single particle reconstruction / cryo EM / Resolution: 3.7 Å | |||||||||
 Authors Authors | Spellmon N / Zimmer J | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Molecular basis for polysaccharide recognition and modulated ATP hydrolysis by the O antigen ABC transporter. Authors: Nicholas Spellmon / Artur Muszyński / Ireneusz Górniak / Jiri Vlach / David Hahn / Parastoo Azadi / Jochen Zimmer /  Abstract: O antigens are ubiquitous protective extensions of lipopolysaccharides in the extracellular leaflet of the Gram-negative outer membrane. Following biosynthesis in the cytosol, the lipid-linked ...O antigens are ubiquitous protective extensions of lipopolysaccharides in the extracellular leaflet of the Gram-negative outer membrane. Following biosynthesis in the cytosol, the lipid-linked polysaccharide is transported to the periplasm by the WzmWzt ABC transporter. Often, O antigen secretion requires the chemical modification of its elongating terminus, which the transporter recognizes via a carbohydrate-binding domain (CBD). Here, using components from A. aeolicus, we identify the O antigen structure with methylated mannose or rhamnose as its cap. Crystal and cryo electron microscopy structures reveal how WzmWzt recognizes this cap between its carbohydrate and nucleotide-binding domains in a nucleotide-free state. ATP binding induces drastic conformational changes of its CBD, terminating interactions with the O antigen. ATPase assays and site directed mutagenesis reveal reduced hydrolytic activity upon O antigen binding, likely to facilitate polymer loading into the ABC transporter. Our results elucidate critical steps in the recognition and translocation of polysaccharides by ABC transporters. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27556.map.gz emd_27556.map.gz | 59.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27556-v30.xml emd-27556-v30.xml emd-27556.xml emd-27556.xml | 17.3 KB 17.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_27556_fsc.xml emd_27556_fsc.xml emd_27556_fsc_2.xml emd_27556_fsc_2.xml emd_27556_fsc_3.xml emd_27556_fsc_3.xml emd_27556_fsc_4.xml emd_27556_fsc_4.xml | 8.9 KB 8.9 KB 8.9 KB 8.9 KB | Display Display Display Display |  FSC data file FSC data file |
| Images |  emd_27556.png emd_27556.png | 67.5 KB | ||
| Filedesc metadata |  emd-27556.cif.gz emd-27556.cif.gz | 6.1 KB | ||
| Others |  emd_27556_half_map_1.map.gz emd_27556_half_map_1.map.gz emd_27556_half_map_2.map.gz emd_27556_half_map_2.map.gz | 59.5 MB 59.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27556 http://ftp.pdbj.org/pub/emdb/structures/EMD-27556 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27556 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27556 | HTTPS FTP |
-Related structure data
| Related structure data |  8dn8MC  8dkuC  8dkyC  8dl0C  8dncC  8dneC  8douC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_27556.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27556.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: half map A
| File | emd_27556_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map B
| File | emd_27556_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : O antigen ABC transporter
| Entire | Name: O antigen ABC transporter |
|---|---|
| Components |
|
-Supramolecule #1: O antigen ABC transporter
| Supramolecule | Name: O antigen ABC transporter / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:   Aquifex aeolicus (bacteria) / Strain: VF5 Aquifex aeolicus (bacteria) / Strain: VF5 |
-Macromolecule #1: ABC transporter
| Macromolecule | Name: ABC transporter / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Aquifex aeolicus (bacteria) / Strain: VF5 Aquifex aeolicus (bacteria) / Strain: VF5 |
| Molecular weight | Theoretical: 46.28441 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGIRVFDVWK KYKYYKKPQD RLKEIIFRKP FHEELWVLKG INLEIEKGEV LGIVGPNGAG KSTLLKVITG VTEPDKGFVE RSGKVVGLL ELGTGFNYEL SGLENIYVNA SLLGLSRREI DEKLESIIEF SELDDFINKP LKTYSSGMIM RLAFSIAIHT E PECFIIDE ...String: MGIRVFDVWK KYKYYKKPQD RLKEIIFRKP FHEELWVLKG INLEIEKGEV LGIVGPNGAG KSTLLKVITG VTEPDKGFVE RSGKVVGLL ELGTGFNYEL SGLENIYVNA SLLGLSRREI DEKLESIIEF SELDDFINKP LKTYSSGMIM RLAFSIAIHT E PECFIIDE ALAVGDAHFQ QKCFRKLKEH KQKGGSIIFV SHDMNAVKIL CDRAILLHKG EIIEEGSPET VTQAYYKLMA SL ENKEGIT FLQNGYGNFK AVIKEVRLKS EHGYTNNFPS GDTLFIELDV EAKEDLQDVV AGILIRDRFG QDIFGINTYL MEK KVELKK GKYLFTFKMP LNLAPGKYTL TVALHKGMDH AQECYHWIDN VCNFEVNGFK KEQFVGVCYL PTEFNYRKIP KLHH HHHH UniProtKB: ABC transporter |
-Macromolecule #2: Transport permease protein
| Macromolecule | Name: Transport permease protein / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Aquifex aeolicus (bacteria) / Strain: VF5 Aquifex aeolicus (bacteria) / Strain: VF5 |
| Molecular weight | Theoretical: 30.027871 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MNLSLILELV RQEIKNRYAD TVLGIWWAFL WPILLVLIYT LIFSHLIGAK LGHENTVYAY SIYLSSGIFP WFFFSNSLSR ITGIFTEKK FLFTKIPIRL EVFPVVVIIS ELINYLIGIS LVTLISFITL GFEGIKYFYL FPVALYLMIV YSFSIGMVLG T LNVFFRDI ...String: MNLSLILELV RQEIKNRYAD TVLGIWWAFL WPILLVLIYT LIFSHLIGAK LGHENTVYAY SIYLSSGIFP WFFFSNSLSR ITGIFTEKK FLFTKIPIRL EVFPVVVIIS ELINYLIGIS LVTLISFITL GFEGIKYFYL FPVALYLMIV YSFSIGMVLG T LNVFFRDI KEIIGVFLQI FFWFTPIVYT LDILPPFVKK LIYYNPMYPV VSIHHLVFVN YLDLHLYSLL GFLLASPLVF FV SYYFFKK LEKDIKDFA UniProtKB: Transport permease protein |
-Macromolecule #3: 3-O-methyl-alpha-D-mannopyranose
| Macromolecule | Name: 3-O-methyl-alpha-D-mannopyranose / type: ligand / ID: 3 / Number of copies: 2 / Formula: U90 |
|---|---|
| Molecular weight | Theoretical: 194.182 Da |
| Chemical component information |  ChemComp-U90: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.0 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 52.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)