[English] 日本語
 Yorodumi
Yorodumi- EMDB-27553: Cryo-EM structure of human Glycine Receptor alpha1-beta heteromer... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of human Glycine Receptor alpha1-beta heteromer, apo state | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | glycine receptor subunit alpha-1 / glycine receptor subunit beta / Green fluorescent protein / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationtaurine binding / negative regulation of transmission of nerve impulse / positive regulation of acrosome reaction / acrosome reaction / synaptic transmission, glycinergic / glycine-gated chloride channel complex / Neurotransmitter receptors and postsynaptic signal transmission / gamma-aminobutyric acid receptor clustering / neuromuscular process controlling posture / postsynaptic specialization ...taurine binding / negative regulation of transmission of nerve impulse / positive regulation of acrosome reaction / acrosome reaction / synaptic transmission, glycinergic / glycine-gated chloride channel complex / Neurotransmitter receptors and postsynaptic signal transmission / gamma-aminobutyric acid receptor clustering / neuromuscular process controlling posture / postsynaptic specialization / extracellularly glycine-gated ion channel activity / righting reflex / regulation of respiratory gaseous exchange by nervous system process / extracellularly glycine-gated chloride channel activity / inhibitory synapse / glycinergic synapse / chloride transport / response to alcohol / adult walking behavior / inhibitory postsynaptic potential / glycine binding / cellular response to zinc ion / startle response / cellular response to ethanol / chloride channel complex / neuropeptide signaling pathway / neuronal action potential / monoatomic ion transport / visual perception / ligand-gated monoatomic ion channel activity involved in regulation of presynaptic membrane potential / chloride transmembrane transport / muscle contraction / bioluminescence / regulation of membrane potential / generation of precursor metabolites and energy / transmitter-gated monoatomic ion channel activity involved in regulation of postsynaptic membrane potential / cellular response to amino acid stimulus / GABA-ergic synapse / transmembrane signaling receptor activity / nervous system development / perikaryon / chemical synaptic transmission / postsynaptic membrane / neuron projection / external side of plasma membrane / intracellular membrane-bounded organelle / neuronal cell body / synapse / dendrite / protein-containing complex binding / zinc ion binding / identical protein binding / membrane / plasma membrane / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.55 Å | |||||||||
 Authors Authors | Liu X / Wang W | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Asymmetric gating of a human hetero-pentameric glycine receptor. Authors: Xiaofen Liu / Weiwei Wang /  Abstract: Hetero-pentameric Cys-loop receptors constitute a major type of neurotransmitter receptors that enable signal transmission and processing in the nervous system. Despite intense investigations into ...Hetero-pentameric Cys-loop receptors constitute a major type of neurotransmitter receptors that enable signal transmission and processing in the nervous system. Despite intense investigations into their working mechanism and pharmaceutical potentials, how neurotransmitters activate these receptors remains unclear due to the lack of high-resolution structural information in the activated open state. Here we report near-atomic resolution structures resolved in digitonin consistent with all principle functional states of the human α1β GlyR, which is a major Cys-loop receptor that mediates inhibitory neurotransmission in the central nervous system of adults. Glycine binding induces cooperative and symmetric structural rearrangements in the neurotransmitter-binding extracellular domain but asymmetrical pore dilation in the transmembrane domain. Symmetric response in the extracellular domain is consistent with electrophysiological data showing cooperative glycine activation and contribution from both α1 and β subunits. A set of functionally essential but differentially charged amino acid residues in the transmembrane domain of the α1 and β subunits explains asymmetric activation. These findings provide a foundation for understanding how the gating of the Cys-loop receptor family members diverges to accommodate specific physiological environments. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27553.map.gz emd_27553.map.gz | 86 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27553-v30.xml emd-27553-v30.xml emd-27553.xml emd-27553.xml | 20.2 KB 20.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_27553.png emd_27553.png | 104.2 KB | ||
| Filedesc metadata |  emd-27553.cif.gz emd-27553.cif.gz | 7 KB | ||
| Others |  emd_27553_half_map_1.map.gz emd_27553_half_map_1.map.gz emd_27553_half_map_2.map.gz emd_27553_half_map_2.map.gz | 84.7 MB 84.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27553 http://ftp.pdbj.org/pub/emdb/structures/EMD-27553 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27553 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27553 | HTTPS FTP |
-Related structure data
| Related structure data |  8dn3MC  8dn2C  8dn4C  8dn5C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_27553.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27553.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_27553_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
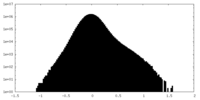
| Projections & Slices |
| ||||||||||||
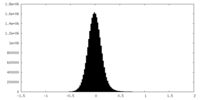
| Density Histograms |
-Half map: #1
| File | emd_27553_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
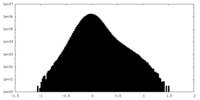
| Projections & Slices |
| ||||||||||||
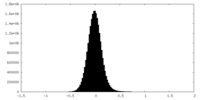
| Density Histograms |
- Sample components
Sample components
+Entire : heteromeric glycine receptor alpha-1 and beta, apo state
+Supramolecule #1: heteromeric glycine receptor alpha-1 and beta, apo state
+Macromolecule #1: Glycine receptor subunit alpha-1
+Macromolecule #2: Glycine receptor subunit beta,Green fluorescent protein,Glycine r...
+Macromolecule #3: 2-acetamido-2-deoxy-beta-D-glucopyranose
+Macromolecule #4: HEXANE
+Macromolecule #5: UNDECANE
+Macromolecule #6: nonane
+Macromolecule #7: N-BUTANE
+Macromolecule #8: HEPTANE
+Macromolecule #9: DECANE
+Macromolecule #10: CHLORIDE ION
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 6 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 400 / Support film - Material: GOLD / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 70 sec. | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 69.6 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Sample stage | Specimen holder model: GATAN LIQUID NITROGEN / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 3.55 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 29850 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)