[English] 日本語
 Yorodumi
Yorodumi- EMDB-27419: Cryo-EM Structure of RSV prefusion F trimer in complex with three... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM Structure of RSV prefusion F trimer in complex with three MxR Fabs | |||||||||
 Map data Map data | Sharp map to 2.2A resolution | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Cross-neutralizing / RSV / HMPV / mAb / IMMUNE SYSTEM | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  Respiratory syncytial virus A2 Respiratory syncytial virus A2 | |||||||||
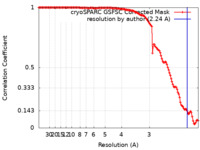
| Method | single particle reconstruction / cryo EM / Resolution: 2.24 Å | |||||||||
 Authors Authors | Rodarte JV / Pancera M | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Cross-protective antibodies against common endemic respiratory viruses. Authors: Madelyn Cabán / Justas V Rodarte / Madeleine Bibby / Matthew D Gray / Justin J Taylor / Marie Pancera / Jim Boonyaratanakornkit /  Abstract: Respiratory syncytial virus (RSV), human metapneumovirus (HMPV), and human parainfluenza virus types one (HPIV1) and three (HPIV3) can cause severe disease and death in immunocompromised patients, ...Respiratory syncytial virus (RSV), human metapneumovirus (HMPV), and human parainfluenza virus types one (HPIV1) and three (HPIV3) can cause severe disease and death in immunocompromised patients, the elderly, and those with underlying lung disease. A protective monoclonal antibody exists for RSV, but clinical use is limited to high-risk infant populations. Hence, therapeutic options for these viruses in vulnerable patient populations are currently limited. Here, we present the discovery, in vitro characterization, and in vivo efficacy testing of two cross-neutralizing monoclonal antibodies, one targeting both HPIV3 and HPIV1 and the other targeting both RSV and HMPV. The 3 × 1 antibody is capable of targeting multiple parainfluenza viruses; the MxR antibody shares features with other previously reported monoclonal antibodies that are capable of neutralizing both RSV and HMPV. We obtained structures using cryo-electron microscopy of these antibodies in complex with their antigens at 3.62 Å resolution for 3 × 1 bound to HPIV3 and at 2.24 Å for MxR bound to RSV, providing a structural basis for in vitro binding and neutralization. Together, a cocktail of 3 × 1 and MxR could have clinical utility in providing broad protection against four of the respiratory viruses that cause significant morbidity and mortality in at-risk individuals. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27419.map.gz emd_27419.map.gz | 204.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27419-v30.xml emd-27419-v30.xml emd-27419.xml emd-27419.xml | 17.1 KB 17.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_27419_fsc.xml emd_27419_fsc.xml | 12.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_27419.png emd_27419.png | 65.7 KB | ||
| Masks |  emd_27419_msk_1.map emd_27419_msk_1.map | 216 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-27419.cif.gz emd-27419.cif.gz | 6.2 KB | ||
| Others |  emd_27419_half_map_1.map.gz emd_27419_half_map_1.map.gz emd_27419_half_map_2.map.gz emd_27419_half_map_2.map.gz | 200.6 MB 200.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27419 http://ftp.pdbj.org/pub/emdb/structures/EMD-27419 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27419 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27419 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_27419.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27419.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharp map to 2.2A resolution | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.02885 Å | ||||||||||||||||||||||||||||||||||||
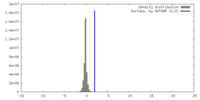
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_27419_msk_1.map emd_27419_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
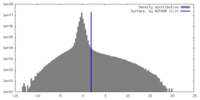
| Density Histograms |
-Half map: Half Map A
| File | emd_27419_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
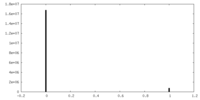
| Density Histograms |
-Half map: Half Map B
| File | emd_27419_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
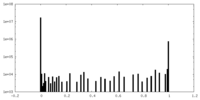
| Density Histograms |
- Sample components
Sample components
-Entire : Trimeric complex of three MxR Fabs bound to RSV preF protomer
| Entire | Name: Trimeric complex of three MxR Fabs bound to RSV preF protomer |
|---|---|
| Components |
|
-Supramolecule #1: Trimeric complex of three MxR Fabs bound to RSV preF protomer
| Supramolecule | Name: Trimeric complex of three MxR Fabs bound to RSV preF protomer type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 339 KDa |
-Macromolecule #1: Fusion glycoprotein F0
| Macromolecule | Name: Fusion glycoprotein F0 / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Respiratory syncytial virus A2 Respiratory syncytial virus A2 |
| Molecular weight | Theoretical: 63.526602 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MELLILKANA ITTILTAVTF CFASGQNITE EFYQSTCSAV SKGYLSALRT GWYTSVITIE LSNIKENKCN GTDAKVKLIK QELDKYKNA VTELQLLMQS TPATNNRARR ELPRFMNYTL NNAKKTNVTL SKKRKRRFLG FLLGVGSAIA SGVAVCKVLH L EGEVNKIK ...String: MELLILKANA ITTILTAVTF CFASGQNITE EFYQSTCSAV SKGYLSALRT GWYTSVITIE LSNIKENKCN GTDAKVKLIK QELDKYKNA VTELQLLMQS TPATNNRARR ELPRFMNYTL NNAKKTNVTL SKKRKRRFLG FLLGVGSAIA SGVAVCKVLH L EGEVNKIK SALLSTNKAV VSLSNGVSVL TFKVLDLKNY IDKQLLPILN KQSCSISNIE TVIEFQQKNN RLLEITREFS VN AGVTTPV STYMLTNSEL LSLINDMPIT NDQKKLMSNN VQIVRQQSYS IMCIIKEEVL AYVVQLPLYG VIDTPCWKLH TSP LCTTNT KEGSNICLTR TDRGWYCDNA GSVSFFPQAE TCKVQSNRVF CDTMNSLTLP SEVNLCNVDI FNPKYDCKIM TSKT DVSSS VITSLGAIVS CYGKTKCTAS NKNRGIIKTF SNGCDYVSNK GVDTVSVGNT LYYVNKQEGK SLYVKGEPII NFYDP LVFP SDEFDASISQ VNEKINQSLA FIRKSDELLS AIGGYIPEAP RDGQAYVRKD GEWVLLSTFL GLNDIFEAQK IEWHEG SHH HHHHHH |
-Macromolecule #2: mAb MxR Heavy Chain, VH region
| Macromolecule | Name: mAb MxR Heavy Chain, VH region / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 12.952548 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EVQVVESGGG LVKPGGSLRL SCAASGFPFS SYKMDWVRQA PGKGLEWVSS ISASGSYINY ADSVKGRFTI SRDNAKNSLY LQMKSLRAD DTAVYFCARD GGRELSPFEK WGQGILVTVS S |
-Macromolecule #3: mAb MxR Light Chain, VL region
| Macromolecule | Name: mAb MxR Light Chain, VL region / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 11.606782 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QSVLTQPPSV SGAPGQRVTI SCTGTNSNIG TGYDVHWYQQ LPGTAPKVVL FDNNNRPSGV PDRFSGSKSG TSAALAITGL QAEDEAVYY CQSYDKSLGG WVFGGGTKLT VL |
-Macromolecule #4: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 4 / Number of copies: 6 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.21 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 295 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)