[English] 日本語
 Yorodumi
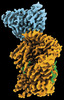
Yorodumi- EMDB-27384: Native serotonin transporter in complex with 15B8 Fab in the pres... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Native serotonin transporter in complex with 15B8 Fab in the presence of methamphetamine | |||||||||
 Map data Map data | the sharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | methamphetamine / SERT / native protein / pig brain / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationserotonin:sodium:chloride symporter activity / antiporter activity / presynapse / neuron projection / endosome membrane / focal adhesion / metal ion binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |   | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | |||||||||
 Authors Authors | Yang D / Gouaux E | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: To be published Journal: To be publishedTitle: Native serotonin transporter in complex with 15B8 Fab in the presence of methamphetamine Authors: Yang D / Gouaux E | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27384.map.gz emd_27384.map.gz | 168 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27384-v30.xml emd-27384-v30.xml emd-27384.xml emd-27384.xml | 21.5 KB 21.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_27384.png emd_27384.png | 113.5 KB | ||
| Filedesc metadata |  emd-27384.cif.gz emd-27384.cif.gz | 6.8 KB | ||
| Others |  emd_27384_additional_1.map.gz emd_27384_additional_1.map.gz emd_27384_half_map_1.map.gz emd_27384_half_map_1.map.gz emd_27384_half_map_2.map.gz emd_27384_half_map_2.map.gz | 88.6 MB 165.1 MB 165.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27384 http://ftp.pdbj.org/pub/emdb/structures/EMD-27384 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27384 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27384 | HTTPS FTP |
-Related structure data
| Related structure data |  8de4MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_27384.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27384.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | the sharpened map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.831 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: the unsharpened map
| File | emd_27384_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | the unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
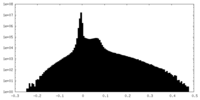
| Density Histograms |
-Half map: half map A
| File | emd_27384_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
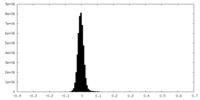
| Density Histograms |
-Half map: half map B
| File | emd_27384_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
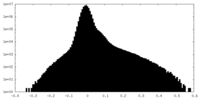
| Density Histograms |
- Sample components
Sample components
-Entire : native serotonin transporter in complex with 15B8 Fab in the pres...
| Entire | Name: native serotonin transporter in complex with 15B8 Fab in the presence of methamphetamine |
|---|---|
| Components |
|
-Supramolecule #1: native serotonin transporter in complex with 15B8 Fab in the pres...
| Supramolecule | Name: native serotonin transporter in complex with 15B8 Fab in the presence of methamphetamine type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Transporter
| Macromolecule | Name: Transporter / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 60.778879 KDa |
| Sequence | String: RETWGKKVDF LLSVIGYAVD LGNVWRFPYI CYQNGGGAFL LPYTIMAIFG GIPLFYMELA LGQYHRNGCI SIWRKICPIF KGIGFAICV IAFYIASYYN TIMAWALYYL ISSFTDQLPW TSCKNSWNTG NCTNYFSEDN VTWMLHSTSP AEEFYTRHVL Q IHRSKGLQ ...String: RETWGKKVDF LLSVIGYAVD LGNVWRFPYI CYQNGGGAFL LPYTIMAIFG GIPLFYMELA LGQYHRNGCI SIWRKICPIF KGIGFAICV IAFYIASYYN TIMAWALYYL ISSFTDQLPW TSCKNSWNTG NCTNYFSEDN VTWMLHSTSP AEEFYTRHVL Q IHRSKGLQ DLGGISWQLA LCIMLIFTII YFSIWKGVKT SGKVVWVTAT FPYIILSILL VRGATLPGAW RGVLFYLKPN WQ KLLETGV WVDAAAQIFF SLGPGFGVLL AFASYNKFNN NCYQDALVTS VVNCMTSFVS GFVIFTVLGY MAEMRNEDVS EVA KDAGPS LLFITYAEAI ANMPASTFFA IIFFLMLITL GLDSTFAGLE GVITAVLDEF PHFWSKRREW LALGVVITCF LGSL ITLTF GGAYVVKLLE EFATGPAVLT VALIEAVAVF WFYGITQFCS DVKEMLGFSP GWFWRICWVA ISPLFLVFII CSFLM SPPQ LWLFQYNYPQ WSIILGYCIG TSSFICIPTY ITYRLIITPG TLKERIVKGI TPETP UniProtKB: Transporter |
-Macromolecule #2: 15B8 Fab heavy chain variable domain
| Macromolecule | Name: 15B8 Fab heavy chain variable domain / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 12.980533 KDa |
| Recombinant expression | Organism:  Baculovirus expression vector pFastBac1-HM Baculovirus expression vector pFastBac1-HM |
| Sequence | String: QVQLQQSGPE LVKLGASVRI SCKASGYRFS YSWMNWVKQR PGKGLEWIGR IYPGDGDTKY SGKFKGKATL TADKSSSTVY MQLSSLTSE DSAVYFCARS AYGSEGFAMD YWGQGTSVT |
-Macromolecule #3: 15B8 Fab light chain variable domain
| Macromolecule | Name: 15B8 Fab light chain variable domain / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 11.776065 KDa |
| Recombinant expression | Organism:  Baculovirus expression vector pFastBac1-HM Baculovirus expression vector pFastBac1-HM |
| Sequence | String: DIVLTQSPAS LAVSLGQRAT ISCRASESVD NYGISFLNWF QQKPGQPPKL LIYAASNQGS GVPARFSGSG SGTYFSLNIH PMEEDDTAV YFCQQTKGVS WTFGGGTKVE I |
-Macromolecule #5: CHLORIDE ION
| Macromolecule | Name: CHLORIDE ION / type: ligand / ID: 5 / Number of copies: 1 / Formula: CL |
|---|---|
| Molecular weight | Theoretical: 35.453 Da |
-Macromolecule #6: SODIUM ION
| Macromolecule | Name: SODIUM ION / type: ligand / ID: 6 / Number of copies: 2 |
|---|---|
| Molecular weight | Theoretical: 22.99 Da |
-Macromolecule #7: (2S)-N-methyl-1-phenylpropan-2-amine
| Macromolecule | Name: (2S)-N-methyl-1-phenylpropan-2-amine / type: ligand / ID: 7 / Number of copies: 1 / Formula: B40 |
|---|---|
| Molecular weight | Theoretical: 149.233 Da |
| Chemical component information | 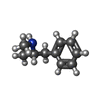 ChemComp-B40: |
-Macromolecule #8: CHOLESTEROL HEMISUCCINATE
| Macromolecule | Name: CHOLESTEROL HEMISUCCINATE / type: ligand / ID: 8 / Number of copies: 2 / Formula: Y01 |
|---|---|
| Molecular weight | Theoretical: 486.726 Da |
| Chemical component information |  ChemComp-Y01: |
-Macromolecule #9: DODECYL-BETA-D-MALTOSIDE
| Macromolecule | Name: DODECYL-BETA-D-MALTOSIDE / type: ligand / ID: 9 / Number of copies: 1 / Formula: LMT |
|---|---|
| Molecular weight | Theoretical: 510.615 Da |
| Chemical component information |  ChemComp-LMT: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)