[English] 日本語
 Yorodumi
Yorodumi- EMDB-27331: The structure of the native cardiac thin filament junction region -
+ Open data
Open data
- Basic information
Basic information
| Entry | 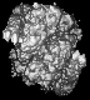 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | The structure of the native cardiac thin filament junction region | ||||||||||||
 Map data Map data | Even half-map | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | thin filament / actin / tropomyosin / troponin / MOTOR PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationStriated Muscle Contraction / troponin C binding / cardiac myofibril / cardiac Troponin complex / troponin complex / actin-myosin filament sliding / regulation of muscle contraction / muscle filament sliding / sarcomere organization / ventricular cardiac muscle tissue morphogenesis ...Striated Muscle Contraction / troponin C binding / cardiac myofibril / cardiac Troponin complex / troponin complex / actin-myosin filament sliding / regulation of muscle contraction / muscle filament sliding / sarcomere organization / ventricular cardiac muscle tissue morphogenesis / heart contraction / myosin binding / tropomyosin binding / troponin I binding / regulation of heart contraction / mesenchyme migration / cardiac muscle contraction / actin filament organization / sarcomere / actin filament / filopodium / response to calcium ion / actin filament binding / actin cytoskeleton / lamellipodium / actin binding / cell body / protein heterodimerization activity / positive regulation of gene expression / protein homodimerization activity / ATP binding / identical protein binding / cytoplasm Similarity search - Function | ||||||||||||
| Biological species |  | ||||||||||||
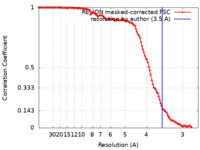
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | ||||||||||||
 Authors Authors | Galkin VE / Risi CM | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: PNAS Nexus / Year: 2023 Journal: PNAS Nexus / Year: 2023Title: High-resolution cryo-EM structure of the junction region of the native cardiac thin filament in relaxed state. Authors: Cristina M Risi / Betty Belknap / Howard D White / Kelly Dryden / Jose R Pinto / P Bryant Chase / Vitold E Galkin /  Abstract: Cardiac contraction depends on molecular interactions among sarcomeric proteins coordinated by the rising and falling intracellular Ca levels. Cardiac thin filament (cTF) consists of two strands ...Cardiac contraction depends on molecular interactions among sarcomeric proteins coordinated by the rising and falling intracellular Ca levels. Cardiac thin filament (cTF) consists of two strands composed of actin, tropomyosin (Tm), and equally spaced troponin (Tn) complexes forming regulatory units. Tn binds Ca to move Tm strand away from myosin-binding sites on actin to enable actomyosin cross-bridges required for force generation. The Tn complex has three subunits-Ca-binding TnC, inhibitory TnI, and Tm-binding TnT. Tm strand is comprised of adjacent Tm molecules that overlap "head-to-tail" along the actin filament. The N-terminus of TnT (e.g., TnT1) binds to the Tm overlap region to form the cTF junction region-the region that connects adjacent regulatory units and confers to cTF internal cooperativity. Numerous studies have predicted interactions among actin, Tm, and TnT1 within the junction region, although a direct structural description of the cTF junction region awaited completion. Here, we report a 3.8 Å resolution cryo-EM structure of the native cTF junction region at (pCa 8) Ca conditions. We provide novel insights into the "head-to-tail" interactions between adjacent Tm molecules and interactions between the Tm junction with F-actin. We demonstrate how TnT1 stabilizes the Tm overlap region via its interactions with the Tm C- and N-termini and actin. Our data show that TnT1 works as a joint that anchors the Tm overlap region to actin, which stabilizes the state of the cTF. Our structure provides insight into the molecular basis of cardiac diseases caused by missense mutations in TnT1. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27331.map.gz emd_27331.map.gz | 3.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27331-v30.xml emd-27331-v30.xml emd-27331.xml emd-27331.xml | 20.3 KB 20.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_27331_fsc.xml emd_27331_fsc.xml | 11.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_27331.png emd_27331.png | 87.4 KB | ||
| Filedesc metadata |  emd-27331.cif.gz emd-27331.cif.gz | 6.3 KB | ||
| Others |  emd_27331_half_map_1.map.gz emd_27331_half_map_1.map.gz emd_27331_half_map_2.map.gz emd_27331_half_map_2.map.gz | 102.1 MB 102.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27331 http://ftp.pdbj.org/pub/emdb/structures/EMD-27331 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27331 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27331 | HTTPS FTP |
-Related structure data
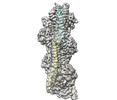
| Related structure data |  8dd0MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_27331.map.gz / Format: CCP4 / Size: 3.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27331.map.gz / Format: CCP4 / Size: 3.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Even half-map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.356 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Even half-map
| File | emd_27331_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Even half-map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Even half-map
| File | emd_27331_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Even half-map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Junction region of the native cardiac thin filament
| Entire | Name: Junction region of the native cardiac thin filament |
|---|---|
| Components |
|
-Supramolecule #1: Junction region of the native cardiac thin filament
| Supramolecule | Name: Junction region of the native cardiac thin filament / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Actin, alpha cardiac muscle 1
| Macromolecule | Name: Actin, alpha cardiac muscle 1 / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 42.064891 KDa |
| Sequence | String: MCDDEETTAL VCDNGSGLVK AGFAGDDAPR AVFPSIVGRP RHQGVMVGMG QKDSYVGDEA QSKRGILTLK YPIEHGIITN WDDMEKIWH HTFYNELRVA PEEHPTLLTE APLNPKANRE KMTQIMFETF NVPAMYVAIQ AVLSLYASGR TTGIVLDSGD G VTHNVPIY ...String: MCDDEETTAL VCDNGSGLVK AGFAGDDAPR AVFPSIVGRP RHQGVMVGMG QKDSYVGDEA QSKRGILTLK YPIEHGIITN WDDMEKIWH HTFYNELRVA PEEHPTLLTE APLNPKANRE KMTQIMFETF NVPAMYVAIQ AVLSLYASGR TTGIVLDSGD G VTHNVPIY EGYALPHAIM RLDLAGRDLT DYLMKILTER GYSFVTTAER EIVRDIKEKL CYVALDFENE MATAASSSSL EK SYELPDG QVITIGNERF RCPETLFQPS FIGMESAGIH ETTYNSIMKC DIDIRKDLYA NNVLSGGTTM YPGIADRMQK EIT ALAPST MKIKIIAPPE RKYSVWIGGS ILASLSTFQQ MWISKQEYDE AGPSIVHRKC F UniProtKB: Actin alpha cardiac muscle 1 |
-Macromolecule #2: Tropomyosin alpha-1 chain
| Macromolecule | Name: Tropomyosin alpha-1 chain / type: protein_or_peptide / ID: 2 / Number of copies: 8 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 32.762656 KDa |
| Sequence | String: MDAIKKKMQM LKLDKENALD RAEQAEADKK AAEDRSKRLE DELVSLQKKL KATEDELDKY SEAPKDAQEK LELAEKKATD AEADVASLN RRIQLVEEEL DRAQERLATA LQKLEEAEKA ADESERGMKV IESRAQKDEE KMEIQEIQLK EAKHIAEDAD R KYEEVARK ...String: MDAIKKKMQM LKLDKENALD RAEQAEADKK AAEDRSKRLE DELVSLQKKL KATEDELDKY SEAPKDAQEK LELAEKKATD AEADVASLN RRIQLVEEEL DRAQERLATA LQKLEEAEKA ADESERGMKV IESRAQKDEE KMEIQEIQLK EAKHIAEDAD R KYEEVARK LVIIESDLER AEERAELSEG KCAELEEELK TVTNNLKSLE AQAEKYSQKE DKYEEEIKVL SDKLKEAETR AE FAERSVT KLEKSIDDLE DELYAQKLKY KAISEELDHA LNDMTSI UniProtKB: Tropomyosin alpha-1 chain |
-Macromolecule #3: Troponin T2, cardiac type
| Macromolecule | Name: Troponin T2, cardiac type / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 35.158918 KDa |
| Sequence | String: MSDVEETVDE YEEEQEEGAA EEEEAWRQDG DEQEEAVEEE AGGEAEAEEA NAEEAGQEED GREAEDGPME ESKPKPRLFM PNLVPPKIP DGERVDFDDI HRKRMKDLNE LQTLIEAHFE NRKKEEELVS LKDRIEKRRA ERAEQQRIRT EREKERQTRL A EERARREE ...String: MSDVEETVDE YEEEQEEGAA EEEEAWRQDG DEQEEAVEEE AGGEAEAEEA NAEEAGQEED GREAEDGPME ESKPKPRLFM PNLVPPKIP DGERVDFDDI HRKRMKDLNE LQTLIEAHFE NRKKEEELVS LKDRIEKRRA ERAEQQRIRT EREKERQTRL A EERARREE EENRRKAEDE ARKKKALSNM MHFGGYIQKQ AQTERKSGKR QTEREKKKKI LAERRKVLAI DHLNEDQLRE KA KELWQSI YNLEAEKFDL QEKFKQQKYE INVLRNRIND NQKVSKTRGK AKVTGRWK UniProtKB: Troponin T, cardiac muscle |
-Macromolecule #4: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 4 / Number of copies: 6 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Macromolecule #5: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 5 / Number of copies: 6 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Grid | Model: EMS Lacey Carbon / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: LACEY / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 15 sec. / Pretreatment - Atmosphere: OTHER |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 5 / Number real images: 24133 / Average electron dose: 34.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.5 µm / Nominal defocus min: 0.5 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)