+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | PI 3-kinase alpha with nanobody 3-126 | ||||||||||||||||||||||||||||||||||||||||||||||||
 Map data Map data | Human PI 3-kinase alpha complex composed of p110alpha and p85alpha with nanobody 3-126 bound, sharpened map | ||||||||||||||||||||||||||||||||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||||||||||||||||||||||||||||||||
 Keywords Keywords | Phosphoinositide 3-kinase (PI3K) / activation / inhibition / nanobody / conformational changes / STRUCTURAL PROTEIN / TRANSFERASE | ||||||||||||||||||||||||||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationperinuclear endoplasmic reticulum membrane / regulation of toll-like receptor 4 signaling pathway / response to muscle inactivity / phosphatidylinositol kinase activity / regulation of actin filament organization / negative regulation of actin filament depolymerization / positive regulation of focal adhesion disassembly / response to butyrate / response to L-leucine / 1-phosphatidylinositol-3-kinase regulator activity ...perinuclear endoplasmic reticulum membrane / regulation of toll-like receptor 4 signaling pathway / response to muscle inactivity / phosphatidylinositol kinase activity / regulation of actin filament organization / negative regulation of actin filament depolymerization / positive regulation of focal adhesion disassembly / response to butyrate / response to L-leucine / 1-phosphatidylinositol-3-kinase regulator activity / phosphatidylinositol 3-kinase regulator activity / IRS-mediated signalling / positive regulation of endoplasmic reticulum unfolded protein response / phosphatidylinositol 3-kinase activator activity / interleukin-18-mediated signaling pathway / T follicular helper cell differentiation / phosphatidylinositol 3-kinase complex / PI3K events in ERBB4 signaling / phosphatidylinositol 3-kinase regulatory subunit binding / myeloid leukocyte migration / autosome genomic imprinting / cellular response to hydrostatic pressure / neurotrophin TRKA receptor binding / regulation of cellular respiration / Activated NTRK2 signals through PI3K / cis-Golgi network / negative regulation of fibroblast apoptotic process / transmembrane receptor protein tyrosine kinase adaptor activity / Activated NTRK3 signals through PI3K / phosphatidylinositol 3-kinase complex, class IB / ErbB-3 class receptor binding / vasculature development / positive regulation of protein localization to membrane / 1-phosphatidylinositol-4-phosphate 3-kinase activity / Signaling by cytosolic FGFR1 fusion mutants / Co-stimulation by ICOS / RHOD GTPase cycle / cardiac muscle cell contraction / phosphatidylinositol 3-kinase complex, class IA / Nephrin family interactions / RHOF GTPase cycle / Signaling by LTK in cancer / anoikis / kinase activator activity / phosphatidylinositol-3-phosphate biosynthetic process / Signaling by LTK / positive regulation of leukocyte migration / MET activates PI3K/AKT signaling / relaxation of cardiac muscle / PI3K/AKT activation / 1-phosphatidylinositol-4,5-bisphosphate 3-kinase activity / negative regulation of stress fiber assembly / RND1 GTPase cycle / RND2 GTPase cycle / phosphatidylinositol-4,5-bisphosphate 3-kinase / RND3 GTPase cycle / positive regulation of filopodium assembly / vascular endothelial growth factor signaling pathway / phosphatidylinositol 3-kinase / growth hormone receptor signaling pathway / insulin binding / Signaling by ALK / 1-phosphatidylinositol-3-kinase activity / RHOV GTPase cycle / Erythropoietin activates Phosphoinositide-3-kinase (PI3K) / PI-3K cascade:FGFR3 / RHOB GTPase cycle / natural killer cell mediated cytotoxicity / GP1b-IX-V activation signalling / negative regulation of macroautophagy / response to dexamethasone / PI-3K cascade:FGFR2 / phosphatidylinositol-mediated signaling / PI-3K cascade:FGFR4 / PI-3K cascade:FGFR1 / RHOC GTPase cycle / RHOJ GTPase cycle / intracellular glucose homeostasis / phosphatidylinositol phosphate biosynthetic process / negative regulation of osteoclast differentiation / Synthesis of PIPs at the plasma membrane / RHOU GTPase cycle / RET signaling / CDC42 GTPase cycle / negative regulation of anoikis / insulin receptor substrate binding / Interleukin-3, Interleukin-5 and GM-CSF signaling / PI3K events in ERBB2 signaling / intercalated disc / PI3K Cascade / T cell differentiation / negative regulation of cell-matrix adhesion / RHOG GTPase cycle / extrinsic apoptotic signaling pathway via death domain receptors / regulation of multicellular organism growth / CD28 dependent PI3K/Akt signaling / Role of LAT2/NTAL/LAB on calcium mobilization / RHOA GTPase cycle / RAC2 GTPase cycle / RAC3 GTPase cycle Similarity search - Function | ||||||||||||||||||||||||||||||||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||||||||||||||||||||||||||||||||||||||
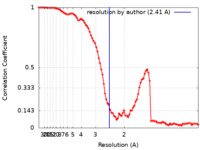
| Method | single particle reconstruction / cryo EM / Resolution: 2.41 Å | ||||||||||||||||||||||||||||||||||||||||||||||||
 Authors Authors | Hart JR / Liu X / Pan C / Liang A / Ueno L / Xu Y / Quezada A / Zou X / Yang S / Zhou Q ...Hart JR / Liu X / Pan C / Liang A / Ueno L / Xu Y / Quezada A / Zou X / Yang S / Zhou Q / Schoonooghe S / Hassanzadeh-Ghassabeh G / Xia T / Shui W / Yang D / Vogt PK / Wang M-W | ||||||||||||||||||||||||||||||||||||||||||||||||
| Funding support |  United States, United States,  China, 15 items China, 15 items
| ||||||||||||||||||||||||||||||||||||||||||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2022 Journal: Proc Natl Acad Sci U S A / Year: 2022Title: Nanobodies and chemical cross-links advance the structural and functional analysis of PI3Kα. Authors: Jonathan R Hart / Xiao Liu / Chen Pan / Anyi Liang / Lynn Ueno / Yingna Xu / Alexandra Quezada / Xinyu Zou / Su Yang / Qingtong Zhou / Steve Schoonooghe / Gholamreza Hassanzadeh-Ghassabeh / ...Authors: Jonathan R Hart / Xiao Liu / Chen Pan / Anyi Liang / Lynn Ueno / Yingna Xu / Alexandra Quezada / Xinyu Zou / Su Yang / Qingtong Zhou / Steve Schoonooghe / Gholamreza Hassanzadeh-Ghassabeh / Tian Xia / Wenqing Shui / Dehua Yang / Peter K Vogt / Ming-Wei Wang /     Abstract: Nanobodies and chemical cross-linking were used to gain information on the identity and positions of flexible domains of PI3Kα. The application of chemical cross-linking mass spectrometry (CXMS) ...Nanobodies and chemical cross-linking were used to gain information on the identity and positions of flexible domains of PI3Kα. The application of chemical cross-linking mass spectrometry (CXMS) facilitated the identification of the p85 domains BH, cSH2, and SH3 as well as their docking positions on the PI3Kα catalytic core. Binding of individual nanobodies to PI3Kα induced activation or inhibition of enzyme activity and caused conformational changes that could be correlated with enzyme function. Binding of nanobody Nb3-126 to the BH domain of p85α substantially improved resolution for parts of the PI3Kα complex, and binding of nanobody Nb3-159 induced a conformation of PI3Kα that is distinct from known PI3Kα structures. The analysis of CXMS data also provided mechanistic insights into the molecular underpinning of the flexibility of PI3Kα. | ||||||||||||||||||||||||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27327.map.gz emd_27327.map.gz | 39.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27327-v30.xml emd-27327-v30.xml emd-27327.xml emd-27327.xml | 21.9 KB 21.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_27327_fsc.xml emd_27327_fsc.xml | 11.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_27327.png emd_27327.png | 48.3 KB | ||
| Masks |  emd_27327_msk_1.map emd_27327_msk_1.map | 42 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-27327.cif.gz emd-27327.cif.gz | 7.2 KB | ||
| Others |  emd_27327_half_map_1.map.gz emd_27327_half_map_1.map.gz emd_27327_half_map_2.map.gz emd_27327_half_map_2.map.gz | 39.1 MB 39.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27327 http://ftp.pdbj.org/pub/emdb/structures/EMD-27327 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27327 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27327 | HTTPS FTP |
-Related structure data
| Related structure data |  8dcpMC  8dcxC  8dd4C  8dd8C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_27327.map.gz / Format: CCP4 / Size: 42 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27327.map.gz / Format: CCP4 / Size: 42 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Human PI 3-kinase alpha complex composed of p110alpha and p85alpha with nanobody 3-126 bound, sharpened map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.535 Å | ||||||||||||||||||||||||||||||||||||
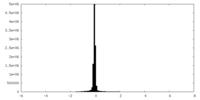
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_27327_msk_1.map emd_27327_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
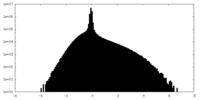
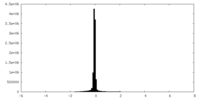
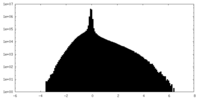
| Density Histograms |
-Half map: Human PI 3-kinase alpha complex composed of p110alpha...
| File | emd_27327_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Human PI 3-kinase alpha complex composed of p110alpha and p85alpha with nanobody 3-126 bound, half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Human PI 3-kinase alpha complex composed of p110alpha...
| File | emd_27327_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Human PI 3-kinase alpha complex composed of p110alpha and p85alpha with nanobody 3-126 bound, half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Human PI 3-kinase alpha complex composed of p110alpha and p85alph...
| Entire | Name: Human PI 3-kinase alpha complex composed of p110alpha and p85alpha with nanobody 3-126 bound |
|---|---|
| Components |
|
-Supramolecule #1: Human PI 3-kinase alpha complex composed of p110alpha and p85alph...
| Supramolecule | Name: Human PI 3-kinase alpha complex composed of p110alpha and p85alpha with nanobody 3-126 bound type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit ...
| Macromolecule | Name: Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: phosphatidylinositol-4,5-bisphosphate 3-kinase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 127.822578 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MSYYHHHHHH DYDIPTTENL YFQGAMGSMP PRPSSGELWG IHLMPPRILV ECLLPNGMIV TLECLREATL ITIKHELFKE ARKYPLHQL LQDESSYIFV SVTQEAEREE FFDETRRLCD LRLFQPFLKV IEPVGNREEK ILNREIGFAI GMPVCEFDMV K DPEVQDFR ...String: MSYYHHHHHH DYDIPTTENL YFQGAMGSMP PRPSSGELWG IHLMPPRILV ECLLPNGMIV TLECLREATL ITIKHELFKE ARKYPLHQL LQDESSYIFV SVTQEAEREE FFDETRRLCD LRLFQPFLKV IEPVGNREEK ILNREIGFAI GMPVCEFDMV K DPEVQDFR RNILNVCKEA VDLRDLNSPH SRAMYVYPPN VESSPELPKH IYNKLDKGQI IVVIWVIVSP NNDKQKYTLK IN HDCVPEQ VIAEAIRKKT RSMLLSSEQL KLCVLEYQGK YILKVCGCDE YFLEKYPLSQ YKYIRSCIML GRMPNLMLMA KES LYSQLP MDCFTMPSYS RRISTATPYM NGETSTKSLW VINSALRIKI LCATYVNVNI RDIDKIYVRT GIYHGGEPLC DNVN TQRVP CSNPRWNEWL NYDIYIPDLP RAARLCLSIC SVKGRKGAKE EHCPLAWGNI NLFDYTDTLV SGKMALNLWP VPHGL EDLL NPIGVTGSNP NKETPCLELE FDWFSSVVKF PDMSVIEEHA NWSVSREAGF SYSHAGLSNR LARDNELREN DKEQLK AIS TRDPLSEITE QEKDFLWSHR HYCVTIPEIL PKLLLSVKWN SRDEVAQMYC LVKDWPPIKP EQAMELLDCN YPDPMVR GF AVRCLEKYLT DDKLSQYLIQ LVQVLKYEQY LDNLLVRFLL KKALTNQRIG HFFFWHLKSE MHNKTVSQRF GLLLESYC R ACGMYLKHLN RQVEAMEKLI NLTDILKQEK KDETQKVQMK FLVEQMRRPD FMDALQGFLS PLNPAHQLGN LRLEECRIM SSAKRPLWLN WENPDIMSEL LFQNNEIIFK NGDDLRQDML TLQIIRIMEN IWQNQGLDLR MLPYGCLSIG DCVGLIEVVR NSHTIMQIQ CKGGLKGALQ FNSHTLHQWL KDKNKGEIYD AAIDLFTRSC AGYCVATFIL GIGDRHNSNI MVKDDGQLFH I DFGHFLDH KKKKFGYKRE RVPFVLTQDF LIVISKGAQE CTKTREFERF QEMCYKAYLA IRQHANLFIN LFSMMLGSGM PE LQSFDDI AYIRKTLALD KTEQEALEYF MKQMNDAHHG GWTTKMDWIF HTIKQHALN UniProtKB: Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform |
-Macromolecule #2: Phosphatidylinositol 3-kinase regulatory subunit alpha
| Macromolecule | Name: Phosphatidylinositol 3-kinase regulatory subunit alpha type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 83.623203 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MAEGYQYRAL YDYKKEREED IDLHLGDILT VNKGSLVALG FSDGQEARPE EIGWLNGYNE TTGERGDFPG TYVEYIGRKK ISPPTPKPR PPRPLPVAPG SSKTEADVEQ QALTLPDLAE QFAPPDIAPP LLIKLVEAIE KKGLECSTLY RTQSSSNLAE L RQLLDCDT ...String: MAEGYQYRAL YDYKKEREED IDLHLGDILT VNKGSLVALG FSDGQEARPE EIGWLNGYNE TTGERGDFPG TYVEYIGRKK ISPPTPKPR PPRPLPVAPG SSKTEADVEQ QALTLPDLAE QFAPPDIAPP LLIKLVEAIE KKGLECSTLY RTQSSSNLAE L RQLLDCDT PSVDLEMIDV HVLADAFKRY LLDLPNPVIP AAVYSEMISL APEVQSSEEY IQLLKKLIRS PSIPHQYWLT LQ YLLKHFF KLSQTSSKNL LNARVLSEIF SPMLFRFSAA SSDNTENLIK VIEILISTEW NERQPAPALP PKPPKPTTVA NNG MNNNMS LQDAEWYWGD ISREEVNEKL RDTADGTFLV RDASTKMHGD YTLTLRKGGN NKLIKIFHRD GKYGFSDPLT FSSV VELIN HYRNESLAQY NPKLDVKLLY PVSKYQQDQV VKEDNIEAVG KKLHEYNTQF QEKSREYDRL YEEYTRTSQE IQMKR TAIE AFNETIKIFE EQCQTQERYS KEYIEKFKRE GNEKEIQRIM HNYDKLKSRI SEIIDSRRRL EEDLKKQAAE YREIDK RMN SIKPDLIQLR KTRDQYLMWL TQKGVRQKKL NEWLGNENTE DQYSLVEDDE DLPHHDEKTW NVGSSNRNKA ENLLRGK RD GTFLVRESSK QGCYACSVVV DGEVKHCVIN KTATGYGFAE PYNLYSSLKE LVLHYQHTSL VQHNDSLNVT LAYPVYAQ Q RR UniProtKB: Phosphatidylinositol 3-kinase regulatory subunit alpha |
-Macromolecule #3: water
| Macromolecule | Name: water / type: ligand / ID: 3 / Number of copies: 21 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.0 mg/mL |
|---|---|
| Buffer | pH: 7.6 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 70.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source: OTHER |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


























 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)