+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of CasLambda (Cas12l) bound to crRNA and DNA | |||||||||
 Map data Map data | LocSpiral map from cryoSPARC half maps | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | CRISPR / RNA Binding Protein / DNA binding protein / phage / viral protein / enzyme / ribonucleoprotein / RNA BINDING PROTEIN-RNA-DNA complex | |||||||||
| Biological species |  uncultured virus (environmental samples) uncultured virus (environmental samples) | |||||||||
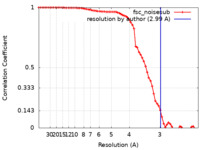
| Method | single particle reconstruction / cryo EM / Resolution: 2.99 Å | |||||||||
 Authors Authors | Al-Shayeb B / Skopintsev P / Soczek K / Doudna J | |||||||||
| Funding support |  United States, United States,  Switzerland, 2 items Switzerland, 2 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2022 Journal: Cell / Year: 2022Title: Diverse virus-encoded CRISPR-Cas systems include streamlined genome editors. Authors: Basem Al-Shayeb / Petr Skopintsev / Katarzyna M Soczek / Elizabeth C Stahl / Zheng Li / Evan Groover / Dylan Smock / Amy R Eggers / Patrick Pausch / Brady F Cress / Carolyn J Huang / Brian ...Authors: Basem Al-Shayeb / Petr Skopintsev / Katarzyna M Soczek / Elizabeth C Stahl / Zheng Li / Evan Groover / Dylan Smock / Amy R Eggers / Patrick Pausch / Brady F Cress / Carolyn J Huang / Brian Staskawicz / David F Savage / Steven E Jacobsen / Jillian F Banfield / Jennifer A Doudna /   Abstract: CRISPR-Cas systems are host-encoded pathways that protect microbes from viral infection using an adaptive RNA-guided mechanism. Using genome-resolved metagenomics, we find that CRISPR systems are ...CRISPR-Cas systems are host-encoded pathways that protect microbes from viral infection using an adaptive RNA-guided mechanism. Using genome-resolved metagenomics, we find that CRISPR systems are also encoded in diverse bacteriophages, where they occur as divergent and hypercompact anti-viral systems. Bacteriophage-encoded CRISPR systems belong to all six known CRISPR-Cas types, though some lack crucial components, suggesting alternate functional roles or host complementation. We describe multiple new Cas9-like proteins and 44 families related to type V CRISPR-Cas systems, including the Casλ RNA-guided nuclease family. Among the most divergent of the new enzymes identified, Casλ recognizes double-stranded DNA using a uniquely structured CRISPR RNA (crRNA). The Casλ-RNA-DNA structure determined by cryoelectron microscopy reveals a compact bilobed architecture capable of inducing genome editing in mammalian, Arabidopsis, and hexaploid wheat cells. These findings reveal a new source of CRISPR-Cas enzymes in phages and highlight their value as genome editors in plant and human cells. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27320.map.gz emd_27320.map.gz | 1.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27320-v30.xml emd-27320-v30.xml emd-27320.xml emd-27320.xml | 23.3 KB 23.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_27320_fsc.xml emd_27320_fsc.xml | 7.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_27320.png emd_27320.png | 84.4 KB | ||
| Masks |  emd_27320_msk_1.map emd_27320_msk_1.map | 30.5 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-27320.cif.gz emd-27320.cif.gz | 6.6 KB | ||
| Others |  emd_27320_additional_1.map.gz emd_27320_additional_1.map.gz emd_27320_additional_2.map.gz emd_27320_additional_2.map.gz emd_27320_half_map_1.map.gz emd_27320_half_map_1.map.gz emd_27320_half_map_2.map.gz emd_27320_half_map_2.map.gz | 28.8 MB 15.4 MB 28.4 MB 28.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27320 http://ftp.pdbj.org/pub/emdb/structures/EMD-27320 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27320 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27320 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_27320.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27320.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | LocSpiral map from cryoSPARC half maps | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.115 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_27320_msk_1.map emd_27320_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: cryoSPARC sharp map
| File | emd_27320_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | cryoSPARC sharp map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: cryoSPARC map
| File | emd_27320_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | cryoSPARC map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_27320_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_27320_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : CasLambda-crRNA-dsDNA ternary structure
| Entire | Name: CasLambda-crRNA-dsDNA ternary structure |
|---|---|
| Components |
|
-Supramolecule #1: CasLambda-crRNA-dsDNA ternary structure
| Supramolecule | Name: CasLambda-crRNA-dsDNA ternary structure / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  uncultured virus (environmental samples) uncultured virus (environmental samples) |
-Macromolecule #1: CasLambda
| Macromolecule | Name: CasLambda / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  uncultured virus (environmental samples) uncultured virus (environmental samples) |
| Molecular weight | Theoretical: 87.337969 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASHKKTESN QIIKTFSFKI KNANGLSLDV LNDAITEYQN YYNICSDWIK DHLTMKISEL YKYIPNEKKN SGYALTLISD EWKDKPMYM MFKKGYPANN RDNAIYETLN TCNTEHYTGN ILNFSDTYYR RFGYVASAIS NYVTKISKMS TGSRSKNISN D SDVDTIME ...String: MASHKKTESN QIIKTFSFKI KNANGLSLDV LNDAITEYQN YYNICSDWIK DHLTMKISEL YKYIPNEKKN SGYALTLISD EWKDKPMYM MFKKGYPANN RDNAIYETLN TCNTEHYTGN ILNFSDTYYR RFGYVASAIS NYVTKISKMS TGSRSKNISN D SDVDTIME QVIYEMEHNG WTSVKDWENQ MEYLESKTDS NPNFVYRMTT LYEFYKSHID EVNSKMETMS IDSLIKFGGC RR KDSKKSM YIMGGSNTPF DITQIGGNSL NIKFSKNLNV DVFGRYDVIK DNTLLVDIIN GHGASFVLKI INDEIYIDIN VSV PFDKKI ATTNKVVGID VNIKHMLLAT NILDDGNVKG YVNIYKEVIN DSDFKKVCNS TVMQYFTDFS KFVTFCPLEF DFLF SRVCN QKGIYNDNSA MEKSFSDVLN KLKWNFIETG DNTKRIYIEN VMKLRSQMKA YAIVKNAYYK QQSEYDFGKS EEFIQ EHPF SNTDKGIEIL NKLDNISKKI LGCRNNIIQY SYNLFEINGY DMVSLEKLTS SQFKKKPFPT VNSLLKYHKI LGCTQE EME KKDIYSVIKK GYYDIIFDND VVTDAKLSAK GELSKFKDDF FNLMIKSIHF ADIKDYFITL SNNGTAGVSL VPSYFTS QM DSIDHKIYFV QDNKSGKLKL ANKHKVRSSQ EKHINGLNAD YNAARNIAYI MENTDCRNMF MKQSRTDKSL YNKPSYET F IKTQGSAVAK LKKEGFVKIL DEASVGSSGH HHHHH |
-Macromolecule #2: RNA (51-MER)
| Macromolecule | Name: RNA (51-MER) / type: rna / ID: 2 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  uncultured virus (environmental samples) uncultured virus (environmental samples) |
| Molecular weight | Theoretical: 16.483719 KDa |
| Sequence | String: AUUGUUGUAA CUCUUAUUUU GUAUGGAGUA AACAACUAGC AUCACCUUCA CC |
-Macromolecule #3: DNA TS
| Macromolecule | Name: DNA TS / type: dna / ID: 3 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  uncultured virus (environmental samples) uncultured virus (environmental samples) |
| Molecular weight | Theoretical: 14.296228 KDa |
| Sequence | String: (DC)(DA)(DT)(DT)(DA)(DA)(DC)(DA)(DT)(DT) (DA)(DC)(DT)(DA)(DA)(DG)(DA)(DG)(DG)(DG) (DT)(DG)(DA)(DA)(DG)(DG)(DT)(DG)(DA) (DT)(DG)(DC)(DT)(DA)(DC)(DA)(DA)(DA)(DC) (DG) (DG)(DT)(DC)(DA)(DA)(DG) |
-Macromolecule #4: DNA NTS
| Macromolecule | Name: DNA NTS / type: dna / ID: 4 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  uncultured virus (environmental samples) uncultured virus (environmental samples) |
| Molecular weight | Theoretical: 14.371228 KDa |
| Sequence | String: (DC)(DT)(DT)(DG)(DA)(DC)(DC)(DG)(DT)(DT) (DT)(DG)(DA)(DT)(DC)(DG)(DT)(DA)(DG)(DT) (DG)(DG)(DA)(DA)(DG)(DT)(DG)(DG)(DG) (DA)(DG)(DA)(DT)(DA)(DG)(DT)(DA)(DA)(DT) (DG) (DT)(DT)(DA)(DA)(DT)(DG) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Support film - Material: GOLD / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 281 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.8 µm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: OTHER |
|---|---|
| Output model |  PDB-8dc2: |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)