[English] 日本語
 Yorodumi
Yorodumi- EMDB-27251: Yeast mitochondrial small subunit assembly intermediate (State 3) -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Yeast mitochondrial small subunit assembly intermediate (State 3) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Ribonucleoprotein complex Mitochondria Biogenesis / RIBOSOME | |||||||||
| Function / homology |  Function and homology information Function and homology informationBranched-chain amino acid catabolism / mitochondrial small ribosomal subunit assembly / 3-hydroxyisobutyryl-CoA hydrolase / 3-hydroxyisobutyryl-CoA hydrolase activity / mitochondrial translational initiation / L-valine catabolic process / Mitochondrial protein degradation / mitochondrial ribosome / mitochondrial small ribosomal subunit / mitochondrial translation ...Branched-chain amino acid catabolism / mitochondrial small ribosomal subunit assembly / 3-hydroxyisobutyryl-CoA hydrolase / 3-hydroxyisobutyryl-CoA hydrolase activity / mitochondrial translational initiation / L-valine catabolic process / Mitochondrial protein degradation / mitochondrial ribosome / mitochondrial small ribosomal subunit / mitochondrial translation / sporulation resulting in formation of a cellular spore / superoxide dismutase activity / methyltransferase activity / peroxisome / 4 iron, 4 sulfur cluster binding / ribosomal small subunit biogenesis / ribosomal small subunit assembly / small ribosomal subunit / small ribosomal subunit rRNA binding / mitochondrial inner membrane / rRNA binding / structural constituent of ribosome / ribosome / translation / mRNA binding / GTP binding / mitochondrion / RNA binding / ATP binding / metal ion binding / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
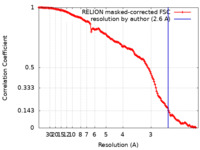
| Method | single particle reconstruction / cryo EM / Resolution: 2.6 Å | |||||||||
 Authors Authors | Burnside C / Harper N / Klinge S | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2023 Journal: Nature / Year: 2023Title: Principles of mitoribosomal small subunit assembly in eukaryotes. Authors: Nathan J Harper / Chloe Burnside / Sebastian Klinge /  Abstract: Mitochondrial ribosomes (mitoribosomes) synthesize proteins encoded within the mitochondrial genome that are assembled into oxidative phosphorylation complexes. Thus, mitoribosome biogenesis is ...Mitochondrial ribosomes (mitoribosomes) synthesize proteins encoded within the mitochondrial genome that are assembled into oxidative phosphorylation complexes. Thus, mitoribosome biogenesis is essential for ATP production and cellular metabolism. Here we used cryo-electron microscopy to determine nine structures of native yeast and human mitoribosomal small subunit assembly intermediates, illuminating the mechanistic basis for how GTPases are used to control early steps of decoding centre formation, how initial rRNA folding and processing events are mediated, and how mitoribosomal proteins have active roles during assembly. Furthermore, this series of intermediates from two species with divergent mitoribosomal architecture uncovers both conserved principles and species-specific adaptations that govern the maturation of mitoribosomal small subunits in eukaryotes. By revealing the dynamic interplay between assembly factors, mitoribosomal proteins and rRNA that are required to generate functional subunits, our structural analysis provides a vignette for how molecular complexity and diversity can evolve in large ribonucleoprotein assemblies. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27251.map.gz emd_27251.map.gz | 228.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27251-v30.xml emd-27251-v30.xml emd-27251.xml emd-27251.xml | 58.1 KB 58.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_27251_fsc.xml emd_27251_fsc.xml | 14.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_27251.png emd_27251.png | 51.2 KB | ||
| Masks |  emd_27251_msk_1.map emd_27251_msk_1.map | 244.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-27251.cif.gz emd-27251.cif.gz | 13.6 KB | ||
| Others |  emd_27251_half_map_1.map.gz emd_27251_half_map_1.map.gz emd_27251_half_map_2.map.gz emd_27251_half_map_2.map.gz | 200.7 MB 200.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27251 http://ftp.pdbj.org/pub/emdb/structures/EMD-27251 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27251 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27251 | HTTPS FTP |
-Related structure data
| Related structure data |  8d8lMC  8cspC  8csqC  8csrC  8cssC  8cstC  8csuC  8d8jC  8d8kC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_27251.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27251.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.057 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_27251_msk_1.map emd_27251_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_27251_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_27251_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : Yeast mitochondrial small subunit assembly intermediate (State 3)
+Supramolecule #1: Yeast mitochondrial small subunit assembly intermediate (State 3)
+Macromolecule #1: Probable S-adenosyl-L-methionine-dependent RNA methyltransferase ...
+Macromolecule #2: 37S ribosomal protein MRP13, mitochondrial
+Macromolecule #3: Ribosomal protein VAR1, mitochondrial
+Macromolecule #4: 37S ribosomal protein SWS2, mitochondrial
+Macromolecule #5: 37S ribosomal protein MRP2, mitochondrial
+Macromolecule #6: 37S ribosomal protein S28, mitochondrial
+Macromolecule #7: 37S ribosomal protein S16, mitochondrial
+Macromolecule #8: 37S ribosomal protein S17, mitochondrial
+Macromolecule #9: 37S ribosomal protein RSM18, mitochondrial
+Macromolecule #10: 37S ribosomal protein S19, mitochondrial
+Macromolecule #11: 37S ribosomal protein MRP21, mitochondrial
+Macromolecule #12: 37S ribosomal protein S25, mitochondrial
+Macromolecule #13: 37S ribosomal protein PET123, mitochondrial
+Macromolecule #14: 37S ribosomal protein S23, mitochondrial
+Macromolecule #15: Mitochondrial 37S ribosomal protein S27
+Macromolecule #16: 37S ribosomal protein S24, mitochondrial
+Macromolecule #17: 37S ribosomal protein MRP10, mitochondrial
+Macromolecule #18: 37S ribosomal protein MRP51, mitochondrial
+Macromolecule #19: 37S ribosomal protein MRP4, mitochondrial
+Macromolecule #20: Protein FYV4, mitochondrial
+Macromolecule #21: 37S ribosomal protein S26, mitochondrial
+Macromolecule #22: 37S ribosomal protein NAM9, mitochondrial
+Macromolecule #23: 37S ribosomal protein MRP1, mitochondrial
+Macromolecule #24: 37S ribosomal protein S5, mitochondrial
+Macromolecule #25: 37S ribosomal protein MRP17, mitochondrial
+Macromolecule #26: 37S ribosomal protein S35, mitochondrial
+Macromolecule #27: 37S ribosomal protein S7, mitochondrial
+Macromolecule #28: 37S ribosomal protein S8, mitochondrial
+Macromolecule #29: 3-hydroxyisobutyryl-CoA hydrolase, mitochondrial
+Macromolecule #30: 37S ribosomal protein S9, mitochondrial
+Macromolecule #32: 37S ribosomal protein S10, mitochondrial
+Macromolecule #33: 37S ribosomal protein S18, mitochondrial
+Macromolecule #34: unknown protein sequence
+Macromolecule #35: 37S ribosomal protein S12, mitochondrial
+Macromolecule #31: 15S ribosomal RNA
+Macromolecule #36: IRON/SULFUR CLUSTER
+Macromolecule #37: ADENOSINE-5'-TRIPHOSPHATE
+Macromolecule #38: MAGNESIUM ION
+Macromolecule #39: water
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 14111 / Average electron dose: 61.73 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 64000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)