+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CIS43 Variant 10 bound to pfCSP Class 4 | |||||||||
 Map data Map data | map | |||||||||
 Sample Sample |
| |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 10.9 Å | |||||||||
 Authors Authors | Gorman J / Kwong PD | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: J Exp Med / Year: 2022 Journal: J Exp Med / Year: 2022Title: Highly protective antimalarial antibodies via precision library generation and yeast display screening. Authors: Bailey B Banach / Prabhanshu Tripathi / Lais Da Silva Pereira / Jason Gorman / Thuy Duong Nguyen / Marlon Dillon / Ahmed S Fahad / Patience K Kiyuka / Bharat Madan / Jacy R Wolfe / Brian ...Authors: Bailey B Banach / Prabhanshu Tripathi / Lais Da Silva Pereira / Jason Gorman / Thuy Duong Nguyen / Marlon Dillon / Ahmed S Fahad / Patience K Kiyuka / Bharat Madan / Jacy R Wolfe / Brian Bonilla / Barbara Flynn / Joseph R Francica / Nicholas K Hurlburt / Neville K Kisalu / Tracy Liu / Li Ou / Reda Rawi / Arne Schön / Chen-Hsiang Shen / I-Ting Teng / Baoshan Zhang / Marie Pancera / Azza H Idris / Robert A Seder / Peter D Kwong / Brandon J DeKosky /   Abstract: The monoclonal antibody CIS43 targets the Plasmodium falciparum circumsporozoite protein (PfCSP) and prevents malaria infection in humans for up to 9 mo following a single intravenous administration. ...The monoclonal antibody CIS43 targets the Plasmodium falciparum circumsporozoite protein (PfCSP) and prevents malaria infection in humans for up to 9 mo following a single intravenous administration. To enhance the potency and clinical utility of CIS43, we used iterative site-saturation mutagenesis and DNA shuffling to screen precise gene-variant yeast display libraries for improved PfCSP antigen recognition. We identified several mutations that improved recognition, predominately in framework regions, and combined these to produce a panel of antibody variants. The most improved antibody, CIS43_Var10, had three mutations and showed approximately sixfold enhanced protective potency in vivo compared to CIS43. Co-crystal and cryo-electron microscopy structures of CIS43_Var10 with the peptide epitope or with PfCSP, respectively, revealed functional roles for each of these mutations. The unbiased site-directed mutagenesis and screening pipeline described here represent a powerful approach to enhance protective potency and to enable broader clinical use of antimalarial antibodies. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27194.map.gz emd_27194.map.gz | 61.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27194-v30.xml emd-27194-v30.xml emd-27194.xml emd-27194.xml | 18.9 KB 18.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_27194.png emd_27194.png | 27.3 KB | ||
| Masks |  emd_27194_msk_1.map emd_27194_msk_1.map | 125 MB |  Mask map Mask map | |
| Others |  emd_27194_half_map_1.map.gz emd_27194_half_map_1.map.gz emd_27194_half_map_2.map.gz emd_27194_half_map_2.map.gz | 115.9 MB 115.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27194 http://ftp.pdbj.org/pub/emdb/structures/EMD-27194 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27194 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27194 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_27194.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27194.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.083 Å | ||||||||||||||||||||||||||||||||||||
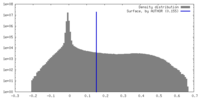
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_27194_msk_1.map emd_27194_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
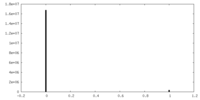
| Density Histograms |
-Half map: half map B
| File | emd_27194_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
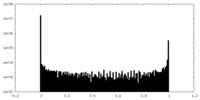
| Density Histograms |
-Half map: half map A
| File | emd_27194_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
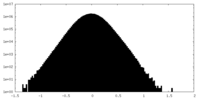
| Density Histograms |
- Sample components
Sample components
-Entire : CIS43 Variant 10 bound to pfCSP Class 4
| Entire | Name: CIS43 Variant 10 bound to pfCSP Class 4 |
|---|---|
| Components |
|
-Supramolecule #1: CIS43 Variant 10 bound to pfCSP Class 4
| Supramolecule | Name: CIS43 Variant 10 bound to pfCSP Class 4 / type: complex / Chimera: Yes / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Var10 Light
| Macromolecule | Name: Var10 Light / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DIVMTQSPDS LAVSLGERAT INCKSSQSVF YSSNNKNYLA WYQQKPGQPP NLLIYWASTR QSGVPDRFSG SGSGTDFTL TISSLQAEDV ASYYCHQYYS SPLTFGGGTK VEIKRTVAAP SVFIFPPSDE QLKSGTASVV C LLNNFYPR EAKVQWKVDN ALQSGNSQES ...String: DIVMTQSPDS LAVSLGERAT INCKSSQSVF YSSNNKNYLA WYQQKPGQPP NLLIYWASTR QSGVPDRFSG SGSGTDFTL TISSLQAEDV ASYYCHQYYS SPLTFGGGTK VEIKRTVAAP SVFIFPPSDE QLKSGTASVV C LLNNFYPR EAKVQWKVDN ALQSGNSQES VTEQDSKDST YSLSSTLTLS KADYEKHKVY ACEVTHQGLS SP VTKSFNR |
-Macromolecule #2: Var10 Heavy
| Macromolecule | Name: Var10 Heavy / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QRQLVQSGAE VKKPGASVKV SCKASGYTFT SYAIHWVRQA PGQRLEWMGW IKAGNGNTRY SQKFQDRVTI TRDTSTTTA YMELSSLRSE DTAVYYCALL TVLTPDDAFD IWGQGTMVTV SSASTKGPSV FPLAPSSKST S GGTAALGC LVKDYFPEPV TVSWNSGALT ...String: QRQLVQSGAE VKKPGASVKV SCKASGYTFT SYAIHWVRQA PGQRLEWMGW IKAGNGNTRY SQKFQDRVTI TRDTSTTTA YMELSSLRSE DTAVYYCALL TVLTPDDAFD IWGQGTMVTV SSASTKGPSV FPLAPSSKST S GGTAALGC LVKDYFPEPV TVSWNSGALT SGVHTFPAVL QSSGLYSLSS VVTVPSSSLG TQTYICNVNH KP SNTKVDK KVEP |
-Macromolecule #3: pfCSPm
| Macromolecule | Name: pfCSPm / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Sequence | String: IATMDSKGSS QKGSRLLLLL VVSNLLLPQG VLAQEYQSYG SSSNTRVLNE LNYDNAGTNL YNELEMNYYG KQENWYSLSS NSASLGENDD GNNEDNEKLR KPKHKKLKQP ADGNPDP NA NPNVDPNANP NVDPNANPNV DPNANPNANP NANPNANPNA NPNANPNANP ...String: IATMDSKGSS QKGSRLLLLL VVSNLLLPQG VLAQEYQSYG SSSNTRVLNE LNYDNAGTNL YNELEMNYYG KQENWYSLSS NSASLGENDD GNNEDNEKLR KPKHKKLKQP ADGNPDP NA NPNVDPNANP NVDPNANPNV DPNANPNANP NANPNANPNA NPNANPNANP NANPNANPNA NPNANPNANP NANPNANPNA NPNANPNANP NVDPNANPNA NPNAN PNAN PNANPNANPN ANPNANPNAN PNANPNANPN ANPNANPNAN PNANPNANPN ANPNANPNKN NQGNGQGHNM PNDPNRNVDE NANANSAVKN NNNEEPSDKH IKEY LNKIQ NSLSTEWSPC SVTCGNGIQV RIKPGSANKP KDELDYANDI EKKICKMEKC SGSGLNDIFE AQKIEWHELE VLFQ |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Details: PBS |
| Grid | Model: C-flat-1.2/1.3 / Support film - Material: CARBON / Support film - topology: HOLEY |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 293 K / Instrument: FEI VITROBOT MARK IV |
| Details | Var10 Fab with pfCSPm |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Average exposure time: 2.0 sec. / Average electron dose: 64.12 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)