+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | Cryo-electron microscopy structure of human kidney Aldehyde Dehydrogenase 1A1 | |||||||||
 マップデータ マップデータ | ||||||||||
 試料 試料 |
| |||||||||
 キーワード キーワード | aldehyde dehydrogenase / ALDH1A1 / HYDROLASE | |||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報fructosamine catabolic process / 3-deoxyglucosone dehydrogenase activity / benzaldehyde dehydrogenase (NAD+) / acetaldehyde dehydrogenase (NAD+) activity / benzaldehyde dehydrogenase (NAD+) activity / aminobutyraldehyde dehydrogenase / retinal dehydrogenase / gamma-aminobutyric acid biosynthetic process / aminobutyraldehyde dehydrogenase (NAD+) activity / maintenance of lens transparency ...fructosamine catabolic process / 3-deoxyglucosone dehydrogenase activity / benzaldehyde dehydrogenase (NAD+) / acetaldehyde dehydrogenase (NAD+) activity / benzaldehyde dehydrogenase (NAD+) activity / aminobutyraldehyde dehydrogenase / retinal dehydrogenase / gamma-aminobutyric acid biosynthetic process / aminobutyraldehyde dehydrogenase (NAD+) activity / maintenance of lens transparency / Fructose catabolism / aldehyde metabolic process / Ethanol oxidation / RA biosynthesis pathway / aldehyde dehydrogenase (NAD+) / androgen binding / cellular detoxification of aldehyde / aldehyde dehydrogenase (NAD+) activity / retinal dehydrogenase (NAD+) activity / retinol metabolic process / negative regulation of cold-induced thermogenesis / retinoid metabolic process / GTPase activator activity / NAD binding / axon / synapse / extracellular exosome / cytoplasm / cytosol 類似検索 - 分子機能 | |||||||||
| 生物種 |  Homo sapiens (ヒト) Homo sapiens (ヒト) | |||||||||
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 2.84 Å | |||||||||
 データ登録者 データ登録者 | Lyu M / Yu EW | |||||||||
| 資金援助 |  米国, 1件 米国, 1件
| |||||||||
 引用 引用 |  ジャーナル: To Be Published ジャーナル: To Be Publishedタイトル: Cryo-electron microscopy structure of human kidney Aldehyde Dehydrogenase 1A1 著者: Lyu M / Yu EW | |||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| 添付画像 |
|---|
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_27176.map.gz emd_27176.map.gz | 97.3 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-27176-v30.xml emd-27176-v30.xml emd-27176.xml emd-27176.xml | 13.5 KB 13.5 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| FSC (解像度算出) |  emd_27176_fsc.xml emd_27176_fsc.xml | 13.6 KB | 表示 |  FSCデータファイル FSCデータファイル |
| 画像 |  emd_27176.png emd_27176.png | 230.8 KB | ||
| Filedesc metadata |  emd-27176.cif.gz emd-27176.cif.gz | 5.4 KB | ||
| その他 |  emd_27176_half_map_1.map.gz emd_27176_half_map_1.map.gz emd_27176_half_map_2.map.gz emd_27176_half_map_2.map.gz | 95.5 MB 95.5 MB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27176 http://ftp.pdbj.org/pub/emdb/structures/EMD-27176 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27176 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27176 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_27176_validation.pdf.gz emd_27176_validation.pdf.gz | 861.4 KB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_27176_full_validation.pdf.gz emd_27176_full_validation.pdf.gz | 860.9 KB | 表示 | |
| XML形式データ |  emd_27176_validation.xml.gz emd_27176_validation.xml.gz | 18.4 KB | 表示 | |
| CIF形式データ |  emd_27176_validation.cif.gz emd_27176_validation.cif.gz | 23.8 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27176 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27176 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27176 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27176 | HTTPS FTP |
-関連構造データ
| 関連構造データ |  8d46MC M: このマップから作成された原子モデル C: 同じ文献を引用 ( |
|---|---|
| 類似構造データ | 類似検索 - 機能・相同性  F&H 検索 F&H 検索 |
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_27176.map.gz / 形式: CCP4 / 大きさ: 103 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_27176.map.gz / 形式: CCP4 / 大きさ: 103 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||
| 密度 |
| ||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
|
-添付データ
-ハーフマップ: #2
| ファイル | emd_27176_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 |
| ||||||||||||
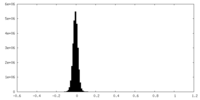
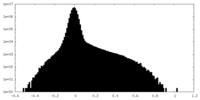
| 密度ヒストグラム |
-ハーフマップ: #1
| ファイル | emd_27176_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 |
| ||||||||||||
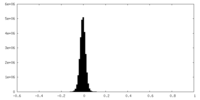
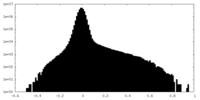
| 密度ヒストグラム |
- 試料の構成要素
試料の構成要素
-全体 : Aldehyde dehydrogenase 1A1
| 全体 | 名称: Aldehyde dehydrogenase 1A1 |
|---|---|
| 要素 |
|
-超分子 #1: Aldehyde dehydrogenase 1A1
| 超分子 | 名称: Aldehyde dehydrogenase 1A1 / タイプ: complex / ID: 1 / 親要素: 0 / 含まれる分子: all |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
-分子 #1: Retinal dehydrogenase 1
| 分子 | 名称: Retinal dehydrogenase 1 / タイプ: protein_or_peptide / ID: 1 / コピー数: 4 / 光学異性体: LEVO EC番号: 酸化還元酵素; アルデヒドまたはケトンに対し酸化酵素として働く; NAD又はNADPを用いる |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 54.924617 KDa |
| 組換発現 | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 配列 | 文字列: MSSSGTPDLP VLLTDLKIQY TKIFINNEWH DSVSGKKFPV FNPATEEELC QVEEGDKEDV DKAVKAARQA FQIGSPWRTM DASERGRLL YKLADLIERD RLLLATMESM NGGKLYSNAY LNDLAGCIKT LRYCAGWADK IQGRTIPIDG NFFTYTRHEP I GVCGQIIP ...文字列: MSSSGTPDLP VLLTDLKIQY TKIFINNEWH DSVSGKKFPV FNPATEEELC QVEEGDKEDV DKAVKAARQA FQIGSPWRTM DASERGRLL YKLADLIERD RLLLATMESM NGGKLYSNAY LNDLAGCIKT LRYCAGWADK IQGRTIPIDG NFFTYTRHEP I GVCGQIIP WNFPLVMLIW KIGPALSCGN TVVVKPAEQT PLTALHVASL IKEAGFPPGV VNIVPGYGPT AGAAISSHMD ID KVAFTGS TEVGKLIKEA AGKSNLKRVT LELGGKSPCI VLADADLDNA VEFAHHGVFY HQGQCCIAAS RIFVEESIYD EFV RRSVER AKKYILGNPL TPGVTQGPQI DKEQYDKILD LIESGKKEGA KLECGGGPWG NKGYFVQPTV FSNVTDEMRI AKEE IFGPV QQIMKFKSLD DVIKRANNTF YGLSAGVFTK DIDKAITISS ALQAGTVWVN CYGVVSAQCP FGGFKMSGNG RELGE YGFH EYTEVKTVTV KISQKNS UniProtKB: Aldehyde dehydrogenase 1A1 |
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 濃度 | .7 mg/mL |
|---|---|
| 緩衝液 | pH: 7.5 |
| グリッド | モデル: Quantifoil R1.2/1.3 / 材質: COPPER / 支持フィルム - 材質: CARBON / 支持フィルム - トポロジー: HOLEY |
| 凍結 | 凍結剤: ETHANE / チャンバー内湿度: 100 % / チャンバー内温度: 277.5 K / 装置: FEI VITROBOT MARK I / 詳細: blot 15 seconds before plunging. |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TITAN KRIOS |
|---|---|
| 撮影 | フィルム・検出器のモデル: GATAN K2 SUMMIT (4k x 4k) 平均電子線量: 39.0 e/Å2 |
| 電子線 | 加速電圧: 300 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | 照射モード: OTHER / 撮影モード: BRIGHT FIELD / 最大 デフォーカス(公称値): 2.5 µm / 最小 デフォーカス(公称値): 1.0 µm |
| 実験機器 |  モデル: Titan Krios / 画像提供: FEI Company |
+ 画像解析
画像解析
-原子モデル構築 1
| 精密化 | プロトコル: AB INITIO MODEL |
|---|---|
| 得られたモデル |  PDB-8d46: |
 ムービー
ムービー コントローラー
コントローラー







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)