[English] 日本語
 Yorodumi
Yorodumi- EMDB-27174: Cryo-electron microscopy structure of human kidney Fructose-bisph... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-electron microscopy structure of human kidney Fructose-bisphosphate aldolase B | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | fructose-bisphosphate aldolase B / ALDOB / HYDROLASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationHereditary fructose intolerance / fructose-1-phosphate aldolase activity / fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate / vacuolar proton-transporting V-type ATPase complex assembly / fructose binding / Fructose catabolism / fructose-bisphosphate aldolase / fructose-bisphosphate aldolase activity / fructose 1,6-bisphosphate metabolic process / fructose metabolic process ...Hereditary fructose intolerance / fructose-1-phosphate aldolase activity / fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate / vacuolar proton-transporting V-type ATPase complex assembly / fructose binding / Fructose catabolism / fructose-bisphosphate aldolase / fructose-bisphosphate aldolase activity / fructose 1,6-bisphosphate metabolic process / fructose metabolic process / Gluconeogenesis / negative regulation of pentose-phosphate shunt / Glycolysis / microtubule organizing center / cytoskeletal protein binding / glycolytic process / gluconeogenesis / centriolar satellite / ATPase binding / molecular adaptor activity / extracellular exosome / identical protein binding / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.8 Å | |||||||||
 Authors Authors | Lyu M / Yu EW | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Cryo-electron microscopy structure of human kidney Fructose-bisphosphate aldolase B Authors: Lyu M / Yu EW | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27174.map.gz emd_27174.map.gz | 97.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27174-v30.xml emd-27174-v30.xml emd-27174.xml emd-27174.xml | 16 KB 16 KB | Display Display |  EMDB header EMDB header |
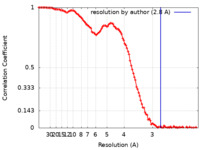
| FSC (resolution estimation) |  emd_27174_fsc.xml emd_27174_fsc.xml | 13.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_27174.png emd_27174.png | 105.2 KB | ||
| Filedesc metadata |  emd-27174.cif.gz emd-27174.cif.gz | 5.7 KB | ||
| Others |  emd_27174_half_map_1.map.gz emd_27174_half_map_1.map.gz emd_27174_half_map_2.map.gz emd_27174_half_map_2.map.gz | 95.3 MB 95.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27174 http://ftp.pdbj.org/pub/emdb/structures/EMD-27174 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27174 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27174 | HTTPS FTP |
-Related structure data
| Related structure data |  8d44MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_27174.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27174.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_27174_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_27174_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Fructose-bisphosphate aldolase B
| Entire | Name: Fructose-bisphosphate aldolase B |
|---|---|
| Components |
|
-Supramolecule #1: Fructose-bisphosphate aldolase B
| Supramolecule | Name: Fructose-bisphosphate aldolase B / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Fructose-bisphosphate aldolase B
| Macromolecule | Name: Fructose-bisphosphate aldolase B / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO / EC number: fructose-bisphosphate aldolase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 39.519969 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAHRFPALTQ EQKKELSEIA QSIVANGKGI LAADESVGTM GNRLQRIKVE NTEENRRQFR EILFSVDSSI NQSIGGVILF HETLYQKDS QGKLFRNILK EKGIVVGIKL DQGGAPLAGT NKETTIQGLD GLSERCAQYK KDGVDFGKWR AVLRIADQCP S SLAIQENA ...String: MAHRFPALTQ EQKKELSEIA QSIVANGKGI LAADESVGTM GNRLQRIKVE NTEENRRQFR EILFSVDSSI NQSIGGVILF HETLYQKDS QGKLFRNILK EKGIVVGIKL DQGGAPLAGT NKETTIQGLD GLSERCAQYK KDGVDFGKWR AVLRIADQCP S SLAIQENA NALARYASIC QQNGLVPIVE PEVIPDGDHD LEHCQYVTEK VLAAVYKALN DHHVYLEGTL LKPNMVTAGH AC TKKYTPE QVAMATVTAL HRTVPAAVPG ICFLSGGMSE EDATLNLNAI NLCPLPKPWK LSFSYGRALQ ASALAAWGGK AAN KEATQE AFMKRAMANC QAAKGQYVHT GSSGAASTQS LFTACYTY UniProtKB: Fructose-bisphosphate aldolase B |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | .7 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: Quantifoil R1.2/1.3 / Support film - Material: CARBON / Support film - topology: HOLEY |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.5 K / Instrument: FEI VITROBOT MARK I / Details: blot 15 seconds before plunging. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 39.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-8d44: |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)