+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human alpha3 Na+/K+-ATPase in its exoplasmic side-open state | |||||||||
 Map data Map data | Human alpha3 Na+/K+-ATPase in its exoplasmic side-open state | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Na/K-ATPase / a3 / b1 / FXYD6 / occluded state / TRANSPORT PROTEIN / TRANSLOCASE-TRANSPORT PROTEIN complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationneuron to neuron synapse / protein transport into plasma membrane raft / Na+/K+-exchanging ATPase / positive regulation of sodium ion export across plasma membrane / positive regulation of potassium ion import across plasma membrane / photoreceptor inner segment membrane / membrane repolarization during cardiac muscle cell action potential / steroid hormone binding / P-type sodium:potassium-exchanging transporter activity / sodium:potassium-exchanging ATPase complex ...neuron to neuron synapse / protein transport into plasma membrane raft / Na+/K+-exchanging ATPase / positive regulation of sodium ion export across plasma membrane / positive regulation of potassium ion import across plasma membrane / photoreceptor inner segment membrane / membrane repolarization during cardiac muscle cell action potential / steroid hormone binding / P-type sodium:potassium-exchanging transporter activity / sodium:potassium-exchanging ATPase complex / membrane repolarization / establishment or maintenance of transmembrane electrochemical gradient / regulation of resting membrane potential / cell communication by electrical coupling involved in cardiac conduction / sodium ion export across plasma membrane / intracellular sodium ion homeostasis / regulation of calcium ion transmembrane transport / relaxation of cardiac muscle / response to glycoside / regulation of cardiac muscle contraction by calcium ion signaling / Basigin interactions / neuronal cell body membrane / cellular response to steroid hormone stimulus / sodium ion transport / organelle membrane / potassium ion import across plasma membrane / intracellular potassium ion homeostasis / ATPase activator activity / Ion transport by P-type ATPases / intercalated disc / lateral plasma membrane / sodium channel regulator activity / sperm flagellum / transporter activator activity / ATP metabolic process / cardiac muscle contraction / Ion homeostasis / photoreceptor inner segment / neuron projection maintenance / T-tubule / sodium ion transmembrane transport / proton transmembrane transport / protein localization to plasma membrane / sarcolemma / caveola / potassium ion transport / cellular response to amyloid-beta / intracellular calcium ion homeostasis / MHC class II protein complex binding / extracellular vesicle / protein-folding chaperone binding / amyloid-beta binding / presynaptic membrane / ATPase binding / regulation of gene expression / protein-macromolecule adaptor activity / basolateral plasma membrane / Potential therapeutics for SARS / postsynaptic membrane / response to hypoxia / cell adhesion / protein stabilization / apical plasma membrane / protein heterodimerization activity / axon / innate immune response / neuronal cell body / synapse / protein kinase binding / glutamatergic synapse / endoplasmic reticulum / Golgi apparatus / ATP hydrolysis activity / extracellular exosome / ATP binding / metal ion binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.9 Å | |||||||||
 Authors Authors | Nguyen PT / Bai X | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Structural basis for gating mechanism of the human sodium-potassium pump. Authors: Phong T Nguyen / Christine Deisl / Michael Fine / Trevor S Tippetts / Emiko Uchikawa / Xiao-Chen Bai / Beth Levine /  Abstract: P2-type ATPase sodium-potassium pumps (Na/K-ATPases) are ion-transporting enzymes that use ATP to transport Na and K on opposite sides of the lipid bilayer against their electrochemical gradients to ...P2-type ATPase sodium-potassium pumps (Na/K-ATPases) are ion-transporting enzymes that use ATP to transport Na and K on opposite sides of the lipid bilayer against their electrochemical gradients to maintain ion concentration gradients across the membranes in all animal cells. Despite the available molecular architecture of the Na/K-ATPases, a complete molecular mechanism by which the Na and K ions access into and are released from the pump remains unknown. Here we report five cryo-electron microscopy (cryo-EM) structures of the human alpha3 Na/K-ATPase in its cytoplasmic side-open (E1), ATP-bound cytoplasmic side-open (E1•ATP), ADP-AlF trapped Na-occluded (E1•P-ADP), BeF trapped exoplasmic side-open (E2P) and MgF trapped K-occluded (E2•P) states. Our work reveals the atomically resolved structural detail of the cytoplasmic gating mechanism of the Na/K-ATPase. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27168.map.gz emd_27168.map.gz | 38 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27168-v30.xml emd-27168-v30.xml emd-27168.xml emd-27168.xml | 19.1 KB 19.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_27168.png emd_27168.png | 43.5 KB | ||
| Filedesc metadata |  emd-27168.cif.gz emd-27168.cif.gz | 6.7 KB | ||
| Others |  emd_27168_half_map_1.map.gz emd_27168_half_map_1.map.gz emd_27168_half_map_2.map.gz emd_27168_half_map_2.map.gz | 31.3 MB 31.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27168 http://ftp.pdbj.org/pub/emdb/structures/EMD-27168 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27168 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27168 | HTTPS FTP |
-Validation report
| Summary document |  emd_27168_validation.pdf.gz emd_27168_validation.pdf.gz | 974.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_27168_full_validation.pdf.gz emd_27168_full_validation.pdf.gz | 974.4 KB | Display | |
| Data in XML |  emd_27168_validation.xml.gz emd_27168_validation.xml.gz | 11.1 KB | Display | |
| Data in CIF |  emd_27168_validation.cif.gz emd_27168_validation.cif.gz | 13 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27168 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27168 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27168 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27168 | HTTPS FTP |
-Related structure data
| Related structure data |  8d3yMC  8d3uC  8d3vC  8d3wC  8d3xC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_27168.map.gz / Format: CCP4 / Size: 40.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27168.map.gz / Format: CCP4 / Size: 40.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Human alpha3 Na+/K+-ATPase in its exoplasmic side-open state | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||
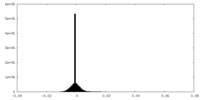
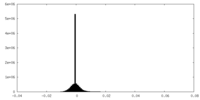
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Human alpha3 Na+/K+-ATPase in its exoplasmic side-open state
| File | emd_27168_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Human alpha3 Na+/K+-ATPase in its exoplasmic side-open state | ||||||||||||
| Projections & Slices |
| ||||||||||||
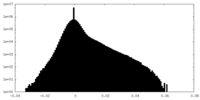
| Density Histograms |
-Half map: Human alpha3 Na+/K+-ATPase in its exoplasmic side-open state
| File | emd_27168_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Human alpha3 Na+/K+-ATPase in its exoplasmic side-open state | ||||||||||||
| Projections & Slices |
| ||||||||||||
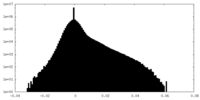
| Density Histograms |
- Sample components
Sample components
-Entire : Na+/K+-ATPase
| Entire | Name: Na+/K+-ATPase |
|---|---|
| Components |
|
-Supramolecule #1: Na+/K+-ATPase
| Supramolecule | Name: Na+/K+-ATPase / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Sodium/potassium-transporting ATPase subunit alpha-3
| Macromolecule | Name: Sodium/potassium-transporting ATPase subunit alpha-3 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: Na+/K+-exchanging ATPase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 111.725195 KDa |
| Recombinant expression | Organism: Mammalian expression vector Flag-MCS-pcDNA3.1 (others) |
| Sequence | String: MGDKKDDKDS PKKNKGKERR DLDDLKKEVA MTEHKMSVEE VCRKYNTDCV QGLTHSKAQE ILARDGPNAL TPPPTTPEWV KFCRQLFGG FSILLWIGAI LCFLAYGIQA GTEDDPSGDN LYLGIVLAAV VIITGCFSYY QEAKSSKIME SFKNMVPQQA L VIREGEKM ...String: MGDKKDDKDS PKKNKGKERR DLDDLKKEVA MTEHKMSVEE VCRKYNTDCV QGLTHSKAQE ILARDGPNAL TPPPTTPEWV KFCRQLFGG FSILLWIGAI LCFLAYGIQA GTEDDPSGDN LYLGIVLAAV VIITGCFSYY QEAKSSKIME SFKNMVPQQA L VIREGEKM QVNAEEVVVG DLVEIKGGDR VPADLRIISA HGCKVDNSSL TGESEPQTRS PDCTHDNPLE TRNITFFSTN CV EGTARGV VVATGDRTVM GRIATLASGL EVGKTPIAIE IEHFIQLITG VAVFLGVSFF ILSLILGYTW LEAVIFLIGI IVA NVPAGL LATVTVCLTL TAKRMARKNC LVKNLEAVET LGSTSTICS(BFD) KTGTLTQNRM TVAHMWFDNQ IHEADTTEDQ SGTSFDKSS HTWVALSHIA GLCNRAVFKG GQDNIPVLKR DVAGDASESA LLKCIELSSG SVKLMRERNK KVAEIPFNST N KYQLSIHE TEDPNDNRYL LVMKGAPERI LDRCSTILLQ GKEQPLDEEM KEAFQNAYLE LGGLGERVLG FCHYYLPEEQ FP KGFAFDC DDVNFTTDNL CFVGLMSMID PPRAAVPDAV GKCRSAGIKV IMVTGDHPIT AKAIAKGVGI ISEGNETVED IAA RLNIPV SQVNPRDAKA CVIHGTDLKD FTSEQIDEIL QNHTEIVFAR TSPQQKLIIV EGCQRQGAIV AVTGDGVNDS PALK KADIG VAMGIAGSDV SKQAADMILL DDNFASIVTG VEEGRLIFDN LKKSIAYTLT SNIPAITPFL LFIMANIPLP LGTIT ILCI ALGTAMVPAI SLAYEAAESD IMKRQPRNPR TDKLVNERLI SMAYGQIGMI QALGGFFSYF VILAENGFLP GNLVGI RLN WDDRTVNDLE DSYGQQWTYE QRKVVEFTCH TAFFVSIVVV QWADLIICKT RRNSVFQQGM KNKILIFGLF EETALAA FL SYCPGMDVAL RMYPLKPSWW FCAFPYSFLI FVYDEIRKLI LRRNPGGWVE KETYY UniProtKB: Sodium/potassium-transporting ATPase subunit alpha-3 |
-Macromolecule #2: Sodium/potassium-transporting ATPase subunit beta-1
| Macromolecule | Name: Sodium/potassium-transporting ATPase subunit beta-1 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 35.108258 KDa |
| Recombinant expression | Organism: Mammalian expression vector Flag-MCS-pcDNA3.1 (others) |
| Sequence | String: MARGKAKEEG SWKKFIWNSE KKEFLGRTGG SWFKILLFYV IFYGCLAGIF IGTIQVMLLT ISEFKPTYQD RVAPPGLTQI PQIQKTEIS FRPNDPKSYE AYVLNIVRFL EKYKDSAQRD DMIFEDCGDV PSEPKERGDF NHERGERKVC RFKLEWLGNC S GLNDETYG ...String: MARGKAKEEG SWKKFIWNSE KKEFLGRTGG SWFKILLFYV IFYGCLAGIF IGTIQVMLLT ISEFKPTYQD RVAPPGLTQI PQIQKTEIS FRPNDPKSYE AYVLNIVRFL EKYKDSAQRD DMIFEDCGDV PSEPKERGDF NHERGERKVC RFKLEWLGNC S GLNDETYG YKEGKPCIII KLNRVLGFKP KPPKNESLET YPVMKYNPNV LPVQCTGKRD EDKDKVGNVE YFGLGNSPGF PL QYYPYYG KLLQPKYLQP LLAVQFTNLT MDTEIRIECK AYGENIGYSE KDRFQGRFDV KIEVKS UniProtKB: Sodium/potassium-transporting ATPase subunit beta-1 |
-Macromolecule #3: FXYD domain-containing ion transport regulator 6
| Macromolecule | Name: FXYD domain-containing ion transport regulator 6 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 10.855645 KDa |
| Recombinant expression | Organism: Mammalian expression vector Flag-MCS-pcDNA3.1 (others) |
| Sequence | String: MATMELVLVF LCSLLAPMVL ASAAEKEKEM DPFHYDYQTL RIGGLVFAVV LFSVGILLIL SRRCKCSFNQ KPRAPGDEEA QVENLITAN ATEPQKAEN UniProtKB: FXYD domain-containing ion transport regulator 6 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 90 sec. |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.6 µm / Nominal defocus min: 1.6 µm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)