+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | VWF tubule derived from dimeric D1-A2 | ||||||||||||
 Map data Map data | Map output from Relion refinement (half maps included) autosharpened in Phenix. | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | VWF / tubule / Blood clotting | ||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
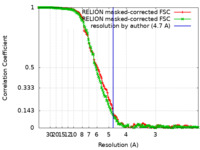
| Method | helical reconstruction / cryo EM / Resolution: 4.7 Å | ||||||||||||
 Authors Authors | Anderson JR / Li J / Springer TA / Brown A | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Blood / Year: 2022 Journal: Blood / Year: 2022Title: Structures of VWF tubules before and after concatemerization reveal a mechanism of disulfide bond exchange. Authors: Jacob R Anderson / Jing Li / Timothy A Springer / Alan Brown /  Abstract: von Willebrand factor (VWF) is an adhesive glycoprotein that circulates in the blood as disulfide-linked concatemers and functions in primary hemostasis. The loss of long VWF concatemers is ...von Willebrand factor (VWF) is an adhesive glycoprotein that circulates in the blood as disulfide-linked concatemers and functions in primary hemostasis. The loss of long VWF concatemers is associated with the excessive bleeding of type 2A von Willebrand disease (VWD). Formation of the disulfide bonds that concatemerize VWF requires VWF to self-associate into helical tubules, yet how the helical tubules template intermolecular disulfide bonds is not known. Here, we report electron cryomicroscopy (cryo-EM) structures of VWF tubules before and after intermolecular disulfide bond formation. The structures provide evidence that VWF tubulates through a charge-neutralization mechanism and that the A1 domain enhances tubule length by crosslinking successive helical turns. In addition, the structures reveal disulfide states before and after disulfide bond-mediated concatemerization. The structures and proposed assembly mechanism provide a foundation to rationalize VWD-causing mutations. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27158.map.gz emd_27158.map.gz | 197.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27158-v30.xml emd-27158-v30.xml emd-27158.xml emd-27158.xml | 18.3 KB 18.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_27158_fsc.xml emd_27158_fsc.xml emd_27158_fsc_2.xml emd_27158_fsc_2.xml | 13.6 KB 13.7 KB | Display Display |  FSC data file FSC data file |
| Images |  emd_27158.png emd_27158.png | 121.6 KB | ||
| Masks |  emd_27158_msk_1.map emd_27158_msk_1.map | 216 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-27158.cif.gz emd-27158.cif.gz | 6.3 KB | ||
| Others |  emd_27158_half_map_1.map.gz emd_27158_half_map_1.map.gz emd_27158_half_map_2.map.gz emd_27158_half_map_2.map.gz | 171.4 MB 171.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27158 http://ftp.pdbj.org/pub/emdb/structures/EMD-27158 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27158 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27158 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_27158.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27158.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map output from Relion refinement (half maps included) autosharpened in Phenix. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_27158_msk_1.map emd_27158_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: 2 of 2 half maps of D1-A2 VWF
| File | emd_27158_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 2 of 2 half maps of D1-A2 VWF | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: 1 of 2 half maps of D1-A2 VWF
| File | emd_27158_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 1 of 2 half maps of D1-A2 VWF | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Von Willebrand Factor tubule derived from dimeric D1-A2
| Entire | Name: Von Willebrand Factor tubule derived from dimeric D1-A2 |
|---|---|
| Components |
|
-Supramolecule #1: Von Willebrand Factor tubule derived from dimeric D1-A2
| Supramolecule | Name: Von Willebrand Factor tubule derived from dimeric D1-A2 type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 14.0 kDa/nm |
-Macromolecule #1: Von Willebrand Factor (D1-A2)
| Macromolecule | Name: Von Willebrand Factor (D1-A2) / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MIPARFAGVL LALALILPGT LCAEGTRGRS STARCSLFGS DFVNTFDGSM YSFAGYCSYL LAGGCQKRSF SIIGDFQNGK RVSLSVYLGE FFDIHLFVNG TVTQGDQRVS MPYASKGLYL ETEAGYYKLS GEAYGFVARI DGSGNFQVLL SDRYFNKTCG LCGNFNIFAE ...String: MIPARFAGVL LALALILPGT LCAEGTRGRS STARCSLFGS DFVNTFDGSM YSFAGYCSYL LAGGCQKRSF SIIGDFQNGK RVSLSVYLGE FFDIHLFVNG TVTQGDQRVS MPYASKGLYL ETEAGYYKLS GEAYGFVARI DGSGNFQVLL SDRYFNKTCG LCGNFNIFAE DDFMTQEGTL TSDPYDFANS WALSSGEQWC ERASPPSSSC NISSGEMQKG LWEQCQLLKS TSVFARCHPL VDPEPFVALC EKTLCECAGG LECACPALLE YARTCAQEGM VLYGWTDHSA CSPVCPAGME YRQCVSPCAR TCQSLHINEM CQERCVDGCS CPEGQLLDEG LCVESTECPC VHSGKRYPPG TSLSRDCNTC ICRNSQWICS NEECPGECLV TGQSHFKSFD NRYFTFSGIC QYLLARDCQD HSFSIVIETV QCADDRDAVC TRSVTVRLPG LHNSLVKLKH GAGVAMDGQD VQLPLLKGDL RIQHTVTASV RLSYGEDLQM DWDGRGRLLV KLSPVYAGKT CGLCGNYNGN QGDDFLTPSG LAEPRVEDFG NAWKLHGDCQ DLQKQHSDPC ALNPRMTRFS EEACAVLTSP TFEACHRAVS PLPYLRNCRY DVCSCSDGRE CLCGALASYA AACAGRGVRV AWREPGRCEL NCPKGQVYLQ CGTPCNLTCR SLSYPDEECN EACLEGCFCP PGLYMDERGD CVPKAQCPCY YDGEIFQPED IFSDHHTMCY CEDGFMHCTM SGVPGSLLPD AVLSSPLSHA SASLSCRPPM VKLVCPADNL RAEGLECAKT CQNYDLECMS MGCVSGCLCP PGMVRHENRC VALERCPCFH QGKEYAPGET VKIGCNTCVC RDRKWNCTDH VCDATCSTIG MAHYLTFDGL KYLFPGECQY VLVQDYCGSN PGTFRILVGN KGCSHPSVKC KKRVTILVEG GEIELFDGEV NVKRPMKDET HFEVVESGRY IILLLGKALS VVWDRHLSIS VVLKQTYQEK VCGLCGNFDG IQNNDLTSSN LQVEEDPVDF GNSWKVSSQC ADTRKVPLDS SPATCHNNIM KQTMVDSSCR ILTSDVFQDC NKLVDPEPYL DVCIYDTCSC ESIGDCACFC DTIAAYAHVC AQHGKVVTWR TATLCPQSCE ERNLRENGYE CEWRYNSCAP ACQVTCQHPE PLACPVQCVE GCHAHCPPGK ILDELLQTCV DPEDCPVCEV AGRRFASGKK VTLNPSDPEH CQICHCDVVN LTCEACQEPG GLVVPPTDAP VSPTTLYVED ISEPPLHDFY CSRLLDLVFL LDGSSRLSEA EFEVLKAFVV DMMERLRISQ KWVRVAVVEY HDGSHAYIGL KDRKRPSELR RIASQVKYAG SQVASTSEVL KYTLFQIFSK IDRPEASRIA LLLMASQEPQ RMSRNFVRYV QGLKKKKVIV IPVGIGPHAN LKQIRLIEKQ APENKAFVLS SVDELEQQRD EIVSYLCDLA PEAPPPTLPP DMAQVTVGPG LLGVSTLGPK RNSMVLDVAF VLEGSDKIGE ADFNRSKEFM EEVIQRMDVG QDSIHVTVLQ YSYMVTVEYP FSEAQSKGDI LQRVREIRYQ GGNRTNTGLA LRYLSDHSFL VSQGDREQAP NLVYMVTGNP ASDEIKRLPG DIQVVPIGVG PNANVQELER IGWPNAPILI QDFETLPREA PDLVLQRCCS |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | helical array |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 5.2 Component:
Details: 100 mM Sodium Cacodylate at pH 5.2, 10 mM CaCl2, and 100 mM NaCl. | ||||||||||||
| Grid | Model: Quantifoil R2/1 / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 2 | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 55.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: OTHER / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL |
|---|
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)