[English] 日本語
 Yorodumi
Yorodumi- EMDB-27154: Human mitochondrial DNA polymerase gamma ternary complex with GC ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human mitochondrial DNA polymerase gamma ternary complex with GC basepair | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | DNA-binding protein / DNA polymerase / TRANSFERASE-DNA complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationgamma DNA polymerase complex / mitochondrial chromosome / mitochondrial DNA replication / Strand-asynchronous mitochondrial DNA replication / positive regulation of DNA-directed DNA polymerase activity / DNA replication proofreading / single-stranded DNA 3'-5' DNA exonuclease activity / Hydrolases; Acting on ester bonds; Exodeoxyribonucleases producing 5'-phosphomonoesters / DNA metabolic process / DNA polymerase processivity factor activity ...gamma DNA polymerase complex / mitochondrial chromosome / mitochondrial DNA replication / Strand-asynchronous mitochondrial DNA replication / positive regulation of DNA-directed DNA polymerase activity / DNA replication proofreading / single-stranded DNA 3'-5' DNA exonuclease activity / Hydrolases; Acting on ester bonds; Exodeoxyribonucleases producing 5'-phosphomonoesters / DNA metabolic process / DNA polymerase processivity factor activity / mitochondrial nucleoid / Lyases; Carbon-oxygen lyases; Other carbon-oxygen lyases / 5'-deoxyribose-5-phosphate lyase activity / base-excision repair, gap-filling / DNA polymerase binding / 3'-5' exonuclease activity / Transcriptional activation of mitochondrial biogenesis / base-excision repair / DNA-templated DNA replication / protease binding / double-stranded DNA binding / in utero embryonic development / DNA-directed DNA polymerase / DNA-directed DNA polymerase activity / mitochondrial matrix / intracellular membrane-bounded organelle / chromatin binding / protein-containing complex / mitochondrion / DNA binding / identical protein binding / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / synthetic construct (others) Homo sapiens (human) / synthetic construct (others) | |||||||||
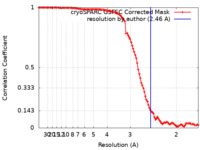
| Method | single particle reconstruction / cryo EM / Resolution: 2.46 Å | |||||||||
 Authors Authors | Park J / Yin YW | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2023 Journal: Nat Struct Mol Biol / Year: 2023Title: Polγ coordinates DNA synthesis and proofreading to ensure mitochondrial genome integrity. Authors: Joon Park / Geoffrey K Herrmann / Patrick G Mitchell / Michael B Sherman / Y Whitney Yin /  Abstract: Accurate replication of mitochondrial DNA (mtDNA) by DNA polymerase γ (Polγ) is essential for maintaining cellular energy supplies, metabolism, and cell cycle control. To illustrate the structural ...Accurate replication of mitochondrial DNA (mtDNA) by DNA polymerase γ (Polγ) is essential for maintaining cellular energy supplies, metabolism, and cell cycle control. To illustrate the structural mechanism for Polγ coordinating polymerase (pol) and exonuclease (exo) activities to ensure rapid and accurate DNA synthesis, we determined four cryo-EM structures of Polγ captured after accurate or erroneous incorporation to a resolution of 2.4-3.0 Å. The structures show that Polγ employs a dual-checkpoint mechanism to sense nucleotide misincorporation and initiate proofreading. The transition from replication to error editing is accompanied by increased dynamics in both DNA and enzyme, in which the polymerase relaxes its processivity and the primer-template DNA unwinds, rotates, and backtracks to shuttle the mismatch-containing primer terminus 32 Å to the exo site for editing. Our structural and functional studies also provide a foundation for analyses of Polγ mutation-induced human diseases and aging. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27154.map.gz emd_27154.map.gz | 58.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27154-v30.xml emd-27154-v30.xml emd-27154.xml emd-27154.xml | 26.3 KB 26.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_27154_fsc.xml emd_27154_fsc.xml | 10.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_27154.png emd_27154.png | 131.7 KB | ||
| Filedesc metadata |  emd-27154.cif.gz emd-27154.cif.gz | 7.8 KB | ||
| Others |  emd_27154_additional_1.map.gz emd_27154_additional_1.map.gz emd_27154_half_map_1.map.gz emd_27154_half_map_1.map.gz emd_27154_half_map_2.map.gz emd_27154_half_map_2.map.gz | 62.5 MB 116.1 MB 116.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27154 http://ftp.pdbj.org/pub/emdb/structures/EMD-27154 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27154 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27154 | HTTPS FTP |
-Validation report
| Summary document |  emd_27154_validation.pdf.gz emd_27154_validation.pdf.gz | 1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_27154_full_validation.pdf.gz emd_27154_full_validation.pdf.gz | 1 MB | Display | |
| Data in XML |  emd_27154_validation.xml.gz emd_27154_validation.xml.gz | 19.1 KB | Display | |
| Data in CIF |  emd_27154_validation.cif.gz emd_27154_validation.cif.gz | 24.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27154 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27154 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27154 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27154 | HTTPS FTP |
-Related structure data
| Related structure data |  8d33MC  8d37C  8d3rC  8d42C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_27154.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27154.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.85 Å | ||||||||||||||||||||||||||||||||||||
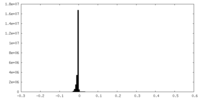
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: CryoSPARC unsharpened map
| File | emd_27154_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoSPARC unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
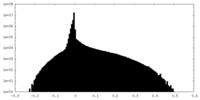
| Density Histograms |
-Half map: #2
| File | emd_27154_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
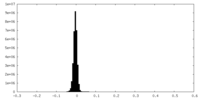
| Density Histograms |
-Half map: #1
| File | emd_27154_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
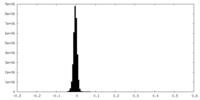
| Density Histograms |
- Sample components
Sample components
-Entire : Human mitochondrial DNA polymerase gamma ternary complex with GC ...
| Entire | Name: Human mitochondrial DNA polymerase gamma ternary complex with GC basepair |
|---|---|
| Components |
|
-Supramolecule #1: Human mitochondrial DNA polymerase gamma ternary complex with GC ...
| Supramolecule | Name: Human mitochondrial DNA polymerase gamma ternary complex with GC basepair type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: DNA polymerase subunit gamma-1
| Macromolecule | Name: DNA polymerase subunit gamma-1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: DNA-directed DNA polymerase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 140.559547 KDa |
| Recombinant expression | Organism: Insect cell expression vector pTIE1 (others) |
| Sequence | String: MSRLLWRKVA GATVGPGPVP APGRWVSSSV PASDPSDGQR RRQQQQQQQQ QQQQQPQQPQ VLSSEGGQLR HNPLDIQMLS RGLHEQIFG QGGEMPGEAA VRRSVEHLQK HGLWGQPAVP LPDVELRLPP LYGDNLDQHF RLLAQKQSLP YLEAANLLLQ A QLPPKPPA ...String: MSRLLWRKVA GATVGPGPVP APGRWVSSSV PASDPSDGQR RRQQQQQQQQ QQQQQPQQPQ VLSSEGGQLR HNPLDIQMLS RGLHEQIFG QGGEMPGEAA VRRSVEHLQK HGLWGQPAVP LPDVELRLPP LYGDNLDQHF RLLAQKQSLP YLEAANLLLQ A QLPPKPPA WAWAEGWTRY GPEGEAVPVA IPEERALVFD VEVCLAEGTC PTLAVAISPS AWYSWCSQRL VEERYSWTSQ LS PADLIPL EVPTGASSPT QRDWQEQLVV GHNVSFDRAH IREQYLIQGS RMRFLDTMSM HMAISGLSSF QRSLWIAAKQ GKH KVQPPT KQGQKSQRKA RRGPAISSWD WLDISSVNSL AEVHRLYVGG PPLEKEPREL FVKGTMKDIR ENFQDLMQYC AQDV WATHE VFQQQLPLFL ERCPHPVTLA GMLEMGVSYL PVNQNWERYL AEAQGTYEEL QREMKKSLMD LANDACQLLS GERYK EDPW LWDLEWDLQE FKQKKAKKVK KEPATASKLP IEGAGAPGDP MDQEDLGPCS EEEEFQQDVM ARACLQKLKG TTELLP KRP QHLPGHPGWY RKLCPRLDDP AWTPGPSLLS LQMRVTPKLM ALTWDGFPLH YSERHGWGYL VPGRRDNLAK LPTGTTL ES AGVVCPYRAI ESLYRKHCLE QGKQQLMPQE AGLAEEFLLT DNSAIWQTVE ELDYLEVEAE AKMENLRAAV PGQPLALT A RGGPKDTQPS YHHGNGPYND VDIPGCWFFK LPHKDGNSCN VGSPFAKDFL PKMEDGTLQA GPGGASGPRA LEINKMISF WRNAHKRISS QMVVWLPRSA LPRAVIRHPD YDEEGLYGAI LPQVVTAGTI TRRAVEPTWL TASNARPDRV GSELKAMVQA PPGYTLVGA DVDSQELWIA AVLGDAHFAG MHGCTAFGWM TLQGRKSRGT DLHSKTATTV GISREHAKIF NYGRIYGAGQ P FAERLLMQ FNHRLTQQEA AEKAQQMYAA TKGLRWYRLS DEGEWLVREL NLPVDRTEGG WISLQDLRKV QRETARKSQW KK WEVVAER AWKGGTESEM FNKLESIATS DIPRTPVLGC CISRALEPSA VQEEFMTSRV NWVVQSSAVD YLHLMLVAMK WLF EEFAID GRFCISIHDE VRYLVREEDR YRAALALQIT NLLTRCMFAY KLGLNDLPQS VAFFSAVDID RCLRKEVTMD CKTP SNPTG MERRYGIPQG EALDIYQIIE LTKGSLEKRS QPGPHHHHHH UniProtKB: DNA polymerase subunit gamma-1 |
-Macromolecule #2: DNA polymerase subunit gamma-2, mitochondrial
| Macromolecule | Name: DNA polymerase subunit gamma-2, mitochondrial / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 55.819867 KDa |
| Recombinant expression | Organism: Insect cell expression vector pTIE1 (others) |
| Sequence | String: MRSRVAVRAC HKVCRCLLSG FGGRVDAGQP ELLTERSSPK GGHVKSHAEL EGNGEHPEAP GSGEGSEALL EICQRRHFLS GSKQQLSRD SLLSGCHPGF GPLGVELRKN LAAEWWTSVV VFREQVFPVD ALHHKPGPLL PGDSAFRLVS AETLREILQD K ELSKEQLV ...String: MRSRVAVRAC HKVCRCLLSG FGGRVDAGQP ELLTERSSPK GGHVKSHAEL EGNGEHPEAP GSGEGSEALL EICQRRHFLS GSKQQLSRD SLLSGCHPGF GPLGVELRKN LAAEWWTSVV VFREQVFPVD ALHHKPGPLL PGDSAFRLVS AETLREILQD K ELSKEQLV AFLENVLKTS GKLRENLLHG ALEHYVNCLD LVNKRLPYGL AQIGVCFHPV FDTKQIRNGV KSIGEKTEAS LV WFTPPRT SNQWLDFWLR HRLQWWRKFA MSPSNFSSSD CQDEEGRKGN KLYYNFPWGK ELIETLWNLG DHELLHMYPG NVS KLHGRD GRKNVVPCVL SVNGDLDRGM LAYLYDSFQL TENSFTRKKN LHRKVLKLHP CLAPIKVALD VGRGPTLELR QVCQ GLFNE LLENGISVWP GYLETMQSSL EQLYSKYDEM SILFTVLVTE TTLENGLIHL RSRDTTMKEM MHISKLKDFL IKYIS SAKN VHHHHHH UniProtKB: DNA polymerase subunit gamma-2 |
-Macromolecule #3: DNA (5'-D(P*AP*AP*AP*AP*CP*GP*AP*CP*GP*GP*CP*CP*AP*GP*TP*GP*CP*CP...
| Macromolecule | Name: DNA (5'-D(P*AP*AP*AP*AP*CP*GP*AP*CP*GP*GP*CP*CP*AP*GP*TP*GP*CP*CP*AP*TP*AP*C)-3') type: dna / ID: 3 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 7.357771 KDa |
| Sequence | String: (DC)(DG)(DA)(DA)(DA)(DA)(DC)(DG)(DA)(DC) (DG)(DG)(DC)(DC)(DA)(DG)(DT)(DG)(DC)(DC) (DA)(DT)(DA)(DC) |
-Macromolecule #4: DNA (25-MER)
| Macromolecule | Name: DNA (25-MER) / type: dna / ID: 4 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 8.644536 KDa |
| Sequence | String: (DC)(DG)(DA)(DG)(DG)(DT)(DA)(DT)(DG)(DG) (DC)(DA)(DC)(DT)(DG)(DG)(DC)(DC)(DG)(DT) (DC)(DG)(DT)(DT)(DT)(DT)(DC)(DG) |
-Macromolecule #5: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 5 / Number of copies: 1 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Macromolecule #6: 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE
| Macromolecule | Name: 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE / type: ligand / ID: 6 / Number of copies: 1 / Formula: DCP |
|---|---|
| Molecular weight | Theoretical: 467.157 Da |
| Chemical component information |  ChemComp-DCP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.2 |
|---|---|
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 200 / Pretreatment - Type: PLASMA CLEANING |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 105000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)