+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
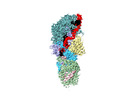
| Title | Cryo-EM structure of guide RNA and target RNA bound Cas7-11 | |||||||||
 Map data Map data | CryoEM structure of typeIIIE Cas7-11 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Crispr / type III-E Cas7-11 / RNA BINDING PROTEIN / RNA BINDING PROTEIN-RNA complex | |||||||||
| Function / homology | : / CRISPR type III-associated protein / RAMP superfamily / defense response to virus / CRISPR-associated RAMP family protein Function and homology information Function and homology information | |||||||||
| Biological species |  Desulfonema ishimotonii (bacteria) Desulfonema ishimotonii (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.82 Å | |||||||||
 Authors Authors | Rai J / Goswami H / Li H | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2022 Journal: Elife / Year: 2022Title: Molecular mechanism of active Cas7-11 in processing CRISPR RNA and interfering target RNA. Authors: Hemant N Goswami / Jay Rai / Anuska Das / Hong Li /  Abstract: Cas7-11 is a Type III-E CRISPR Cas effector that confers programmable RNA cleavage and has potential applications in RNA interference. Cas7-11 encodes a single polypeptide containing four Cas7- and ...Cas7-11 is a Type III-E CRISPR Cas effector that confers programmable RNA cleavage and has potential applications in RNA interference. Cas7-11 encodes a single polypeptide containing four Cas7- and one Cas11-like segments that obscures the distinction between the multi-subunit Class 1 and the single-subunit Class-2 CRISPR Cas systems. We report a cryo-EM (cryo-electron microscopy) structure of the active Cas7-11 from (DiCas7-11) that reveals the molecular basis for RNA processing and interference activities. DiCas7-11 arranges its Cas7- and Cas11-like domains in an extended form that resembles the backbone made up by four Cas7 and one Cas11 subunits in the multi-subunit enzymes. Unlike the multi-subunit enzymes, however, the backbone of DiCas7-11 contains evolutionarily different Cas7 and Cas11 domains, giving rise to their unique functionality. The first Cas7-like domain nearly engulfs the last 15 direct repeat nucleotides in processing and recognition of the CRISPR RNA, and its free-standing fragment retains most of the activity. Both the second and the third Cas7-like domains mediate target RNA cleavage in a metal-dependent manner. The structure and mutational data indicate that the long variable insertion to the fourth Cas7 domain has little impact on RNA processing or targeting, suggesting the possibility for engineering a compact and programmable RNA interference tool. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27138.map.gz emd_27138.map.gz | 96.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27138-v30.xml emd-27138-v30.xml emd-27138.xml emd-27138.xml | 18.8 KB 18.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_27138.png emd_27138.png | 48.4 KB | ||
| Filedesc metadata |  emd-27138.cif.gz emd-27138.cif.gz | 6.9 KB | ||
| Others |  emd_27138_half_map_1.map.gz emd_27138_half_map_1.map.gz emd_27138_half_map_2.map.gz emd_27138_half_map_2.map.gz | 80.9 MB 80.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27138 http://ftp.pdbj.org/pub/emdb/structures/EMD-27138 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27138 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27138 | HTTPS FTP |
-Related structure data
| Related structure data |  8d1vMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_27138.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27138.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoEM structure of typeIIIE Cas7-11 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||
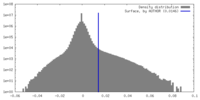
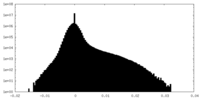
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half-maps1
| File | emd_27138_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-maps1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
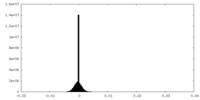
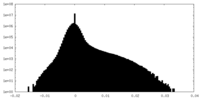
| Density Histograms |
-Half map: Half-map2
| File | emd_27138_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
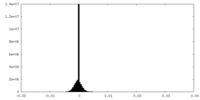
| Density Histograms |
- Sample components
Sample components
-Entire : CryoEM structure of type III-E Cas7-11 from Desulfonema ishimotonii
| Entire | Name: CryoEM structure of type III-E Cas7-11 from Desulfonema ishimotonii |
|---|---|
| Components |
|
-Supramolecule #1: CryoEM structure of type III-E Cas7-11 from Desulfonema ishimotonii
| Supramolecule | Name: CryoEM structure of type III-E Cas7-11 from Desulfonema ishimotonii type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Desulfonema ishimotonii (bacteria) Desulfonema ishimotonii (bacteria) |
-Macromolecule #1: CRISPR-associated RAMP family protein
| Macromolecule | Name: CRISPR-associated RAMP family protein / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Desulfonema ishimotonii (bacteria) Desulfonema ishimotonii (bacteria) |
| Molecular weight | Theoretical: 183.933 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTTTMKISIE FLEPFRMTKW QESTRRNKNN KEFVRGQAFA RWHRNKKDNT KGRPYITGTL LRSAVIRSAE NLLTLSDGKI SEKTCCPGK FDTEDKDRLL QLRQRSTLRW TDKNPCPDNA ETYCPFCELL GRSGNDGKKA EKKDWRFRIH FGNLSLPGKP D FDGPKAIG ...String: MTTTMKISIE FLEPFRMTKW QESTRRNKNN KEFVRGQAFA RWHRNKKDNT KGRPYITGTL LRSAVIRSAE NLLTLSDGKI SEKTCCPGK FDTEDKDRLL QLRQRSTLRW TDKNPCPDNA ETYCPFCELL GRSGNDGKKA EKKDWRFRIH FGNLSLPGKP D FDGPKAIG SQRVLNRVDF KSGKAHDFFK AYEVDHTRFP RFEGEITIDN KVSAEARKLL CDSLKFTDRL CGALCVIRFD EY TPAADSG KQTENVQAEP NANLAEKTAE QIISILDDNK KTEYTRLLAD AIRSLRRSSK LVAGLPKDHD GKDDHYLWDI GKK KKDENS VTIRQILTTS ADTKELKNAG KWREFCEKLG EALYLKSKDM SGGLKITRRI LGDAEFHGKP DRLEKSRSVS IGSV LKETV VCGELVAKTP FFFGAIDEDA KQTDLQVLLT PDNKYRLPRS AVRGILRRDL QTYFDSPCNA ELGGRPCMCK TCRIM RGIT VMDARSEYNA PPEIRHRTRI NPFTGTVAEG ALFNMEVAPE GIVFPFQLRY RGSEDGLPDA LKTVLKWWAE GQAFMS GAA STGKGRFRME NAKYETLDLS DENQRNDYLK NWGWRDEKGL EELKKRLNSG LPEPGNYRDP KWHEINVSIE MASPFIN GD PIRAAVDKRG TDVVTFVKYK AEGEEAKPVC AYKAESFRGV IRSAVARIHM EDGVPLTELT HSDCECLLCQ IFGSEYEA G KIRFEDLVFE SDPEPVTFDH VAIDRFTGGA ADKKKFDDSP LPGSPARPLM LKGSFWIRRD VLEDEEYCKA LGKALADVN NGLYPLGGKS AIGYGQVKSL GIKGDDKRIS RLMNPAFDET DVAVPEKPKT DAEVRIEAEK VYYPHYFVEP HKKVEREEKP CGHQKFHEG RLTGKIRCKL ITKTPLIVPD TSNDDFFRPA DKEARKEKDE YHKSYAFFRL HKQIMIPGSE LRGMVSSVYE T VTNSCFRI FDETKRLSWR MDADHQNVLQ DFLPGRVTAD GKHIQKFSET ARVPFYDKTQ KHFDILDEQE IAGEKPVRMW VK RFIKRLS LVDPAKHPQK KQDNKWKRRK EGIATFIEQK NGSYYFNVVT NNGCTSFHLW HKPDNFDQEK LEGIQNGEKL DCW VRDSRY QKAFQEIPEN DPDGWECKEG YLHVVGPSKV EFSDKKGDVI NNFQGTLPSV PNDWKTIRTN DFKNRKRKNE PVFC CEDDK GNYYTMAKYC ETFFFDLKEN EEYEIPEKAR IKYKELLRVY NNNPQAVPES VFQSRVAREN VEKLKSGDLV YFKHN EKYV EDIVPVRISR TVDDRMIGKR MSADLRPCHG DWVEDGDLSA LNAYPEKRLL LRHPKGLCPA CRLFGTGSYK GRVRFG FAS LENDPEWLIP GKNPGDPFHG GPVMLSLLER PRPTWSIPGS DNKFKVPGRK FYVHHHAWKT IKDGNHPTTG KAIEQSP NN RTVEALAGGN SFSFEIAFEN LKEWELGLLI HSLQLEKGLA HKLGMAKSMG FGSVEIDVES VRLRKDWKQW RNGNSEIP N WLGKGFAKLK EWFRDELDFI ENLKKLLWFP EGDQAPRVCY PMLRKKDDPN GNSGYEELKD GEFKKEDRQK KLTTPWTPW A UniProtKB: CRISPR-associated RAMP family protein |
-Macromolecule #2: CRISPR RNA (34-MER)
| Macromolecule | Name: CRISPR RNA (34-MER) / type: rna / ID: 2 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Desulfonema ishimotonii (bacteria) Desulfonema ishimotonii (bacteria) |
| Molecular weight | Theoretical: 10.837447 KDa |
| Sequence | String: UUGAUGUCAC GGAACACGUU CUUUGAACCA AGCU |
-Macromolecule #3: SS target RNA (5'-R(P*AP*GP*CP*UP*UP*GP*GP*UP*UP*CP*AP*AP*AP*GP*A...
| Macromolecule | Name: SS target RNA (5'-R(P*AP*GP*CP*UP*UP*GP*GP*UP*UP*CP*AP*AP*AP*GP*AP*AP*CP*G)-3') type: rna / ID: 3 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Desulfonema ishimotonii (bacteria) Desulfonema ishimotonii (bacteria) |
| Molecular weight | Theoretical: 5.796516 KDa |
| Sequence | String: AGCUUGGUUC AAAGAACG |
-Macromolecule #4: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 4 / Number of copies: 4 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)