+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | hBest2 1uM Ca2+ (Ca2+-bound) closed state | ||||||||||||
 Map data Map data | phenix.resolve map, resampled | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Ion channel / chloride channel / anion channel / pentamer / TRANSPORT PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationintracellularly ligand-gated monoatomic ion channel activity / ligand-gated monoatomic anion channel activity / bicarbonate channel activity / ligand-gated monoatomic cation channel activity / chloride channel activity / membrane depolarization / chloride channel complex / Stimuli-sensing channels / sensory perception of smell / basolateral plasma membrane ...intracellularly ligand-gated monoatomic ion channel activity / ligand-gated monoatomic anion channel activity / bicarbonate channel activity / ligand-gated monoatomic cation channel activity / chloride channel activity / membrane depolarization / chloride channel complex / Stimuli-sensing channels / sensory perception of smell / basolateral plasma membrane / cilium / metal ion binding / plasma membrane Similarity search - Function | ||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
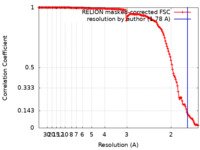
| Method | single particle reconstruction / cryo EM / Resolution: 1.78 Å | ||||||||||||
 Authors Authors | Owji AP / Kittredge A / Hendrickson WA / Tingting Y | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Structures and gating mechanisms of human bestrophin anion channels. Authors: Aaron P Owji / Jiali Wang / Alec Kittredge / Zada Clark / Yu Zhang / Wayne A Hendrickson / Tingting Yang /  Abstract: Bestrophin-1 (Best1) and bestrophin-2 (Best2) are two members of the bestrophin family of calcium (Ca)-activated chloride (Cl) channels with critical involvement in ocular physiology and direct ...Bestrophin-1 (Best1) and bestrophin-2 (Best2) are two members of the bestrophin family of calcium (Ca)-activated chloride (Cl) channels with critical involvement in ocular physiology and direct pathological relevance. Here, we report cryo-EM structures of wild-type human Best1 and Best2 in various states at up to 1.8 Å resolution. Ca-bound Best1 structures illustrate partially open conformations at the two Ca-dependent gates of the channels, in contrast to the fully open conformations observed in Ca-bound Best2, which is in accord with the significantly smaller currents conducted by Best1 in electrophysiological recordings. Comparison of the closed and open states reveals a C-terminal auto-inhibitory segment (AS), which constricts the channel concentrically by wrapping around the channel periphery in an inter-protomer manner and must be released to allow channel opening. Our results demonstrate that removing the AS from Best1 and Best2 results in truncation mutants with similar activities, while swapping the AS between Best1 and Best2 results in chimeric mutants with swapped activities, underlying a key role of the AS in determining paralog specificity among bestrophins. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27127.map.gz emd_27127.map.gz | 70 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27127-v30.xml emd-27127-v30.xml emd-27127.xml emd-27127.xml | 23 KB 23 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_27127_fsc.xml emd_27127_fsc.xml | 14 KB | Display |  FSC data file FSC data file |
| Images |  emd_27127.png emd_27127.png | 104.6 KB | ||
| Filedesc metadata |  emd-27127.cif.gz emd-27127.cif.gz | 6.4 KB | ||
| Others |  emd_27127_additional_1.map.gz emd_27127_additional_1.map.gz emd_27127_half_map_1.map.gz emd_27127_half_map_1.map.gz emd_27127_half_map_2.map.gz emd_27127_half_map_2.map.gz | 122.5 MB 226.6 MB 226.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27127 http://ftp.pdbj.org/pub/emdb/structures/EMD-27127 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27127 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27127 | HTTPS FTP |
-Related structure data
| Related structure data |  8d1eMC  8d1fC  8d1gC  8d1hC  8d1iC  8d1jC  8d1kC  8d1lC  8d1mC  8d1nC  8d1oC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_27127.map.gz / Format: CCP4 / Size: 75.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27127.map.gz / Format: CCP4 / Size: 75.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | phenix.resolve map, resampled | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.40578 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: full map
| File | emd_27127_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | full map | ||||||||||||
| Projections & Slices |
| ||||||||||||
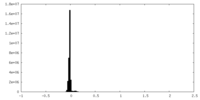
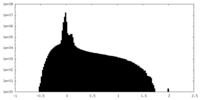
| Density Histograms |
-Half map: CryoSPARC half map A
| File | emd_27127_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoSPARC half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
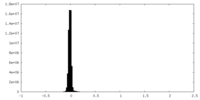
| Density Histograms |
-Half map: CryoSPARC half map B
| File | emd_27127_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoSPARC half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
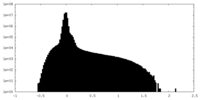
| Density Histograms |
- Sample components
Sample components
-Entire : hBest2 1uM Ca2+ (Ca2+-bound) closed state
| Entire | Name: hBest2 1uM Ca2+ (Ca2+-bound) closed state |
|---|---|
| Components |
|
-Supramolecule #1: hBest2 1uM Ca2+ (Ca2+-bound) closed state
| Supramolecule | Name: hBest2 1uM Ca2+ (Ca2+-bound) closed state / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 234 KDa |
-Macromolecule #1: Bestrophin-2
| Macromolecule | Name: Bestrophin-2 / type: protein_or_peptide / ID: 1 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 46.796898 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MTVTYTARVA NARFGGFSQL LLLWRGSIYK LLWRELLCFL GFYMALSAAY RFVLTEGQKR YFEKLVIYCD QYASLIPVSF VLGFYVTLV VNRWWSQYLC MPLPDALMCV VAGTVHGRDD RGRLYRRTLM RYAGLSAVLI LRSVSTAVFK RFPTIDHVVE A GFMTREER ...String: MTVTYTARVA NARFGGFSQL LLLWRGSIYK LLWRELLCFL GFYMALSAAY RFVLTEGQKR YFEKLVIYCD QYASLIPVSF VLGFYVTLV VNRWWSQYLC MPLPDALMCV VAGTVHGRDD RGRLYRRTLM RYAGLSAVLI LRSVSTAVFK RFPTIDHVVE A GFMTREER KKFENLNSSY NKYWVPCVWF SNLAAQARRE GRIRDNSALK LLLEELNVFR GKCGMLFHYD WISVPLVYTQ VV TIALYSY FLACLIGRQF LDPAQGYKDH DLDLCVPIFT LLQFFFYAGW LKVAEQLINP FGEDDDDFET NFLIDRNFQV SML AVDEMY DDLAVLEKDL YWDAAEARAP YTAATVFQLR QPSFQGSTFD ITLAKEDMQF QRLDGLDGPM GEAPGDFLQR LLPA GAGMV A UniProtKB: Bestrophin-2a |
-Macromolecule #2: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 2 / Number of copies: 5 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Macromolecule #3: 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE
| Macromolecule | Name: 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE / type: ligand / ID: 3 / Number of copies: 50 / Formula: MC3 |
|---|---|
| Molecular weight | Theoretical: 677.933 Da |
| Chemical component information |  ChemComp-MC3: |
-Macromolecule #4: 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13...
| Macromolecule | Name: 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol type: ligand / ID: 4 / Number of copies: 5 / Formula: DU0 |
|---|---|
| Molecular weight | Theoretical: 516.752 Da |
| Chemical component information |  ChemComp-DU0: |
-Macromolecule #5: CHLORIDE ION
| Macromolecule | Name: CHLORIDE ION / type: ligand / ID: 5 / Number of copies: 1 / Formula: CL |
|---|---|
| Molecular weight | Theoretical: 35.453 Da |
-Macromolecule #6: water
| Macromolecule | Name: water / type: ligand / ID: 6 / Number of copies: 629 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.8 Component:
| ||||||||||||
| Grid | Model: UltrAuFoil R0.6/1 / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 20 sec. | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 283.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Specialist optics | Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number real images: 2593 / Average electron dose: 58.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) X (Row.)
X (Row.) Y (Col.)
Y (Col.)