+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Minor conformation of nanodisc embedded ABCA1 | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | sterol transport / ABC transporter / phospholipid transport / MEMBRANE PROTEIN | ||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
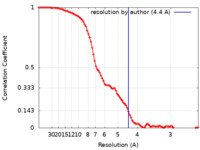
| Method | single particle reconstruction / cryo EM / Resolution: 4.4 Å | ||||||||||||
 Authors Authors | Plummer AM / Culbertson AT / Morales-Perez CL / Liao M | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: J Mol Biol / Year: 2023 Journal: J Mol Biol / Year: 2023Title: Activity and Structural Dynamics of Human ABCA1 in a Lipid Membrane. Authors: Ashlee M Plummer-Medeiros / Alan T Culbertson / Claudio L Morales-Perez / Maofu Liao /   Abstract: The human ATP-binding cassette (ABC) transporter ABCA1 plays a critical role in lipid homeostasis as it extracts sterols and phospholipids from the plasma membrane for excretion to the extracellular ...The human ATP-binding cassette (ABC) transporter ABCA1 plays a critical role in lipid homeostasis as it extracts sterols and phospholipids from the plasma membrane for excretion to the extracellular apolipoprotein A-I and subsequent formation of high-density lipoprotein (HDL) particles. Deleterious mutations of ABCA1 lead to sterol accumulation and are associated with atherosclerosis, poor cardiovascular outcomes, cancer, and Alzheimer's disease. The mechanism by which ABCA1 drives lipid movement is poorly understood, and a unified platform to produce active ABCA1 protein for both functional and structural studies has been missing. In this work, we established a stable expression system for both a human cell-based sterol export assay and protein purification for in vitro biochemical and structural studies. ABCA1 produced in this system was active in sterol export and displayed enhanced ATPase activity after reconstitution into a lipid bilayer. Our single-particle cryo-EM study of ABCA1 in nanodiscs showed protein induced membrane curvature, revealed multiple distinct conformations, and generated a structure of nanodisc-embedded ABCA1 at 4.0-Å resolution representing a previously unknown conformation. Comparison of different ABCA1 structures and molecular dynamics simulations demonstrates both concerted domain movements and conformational variations within each domain. Taken together, our platform for producing and characterizing ABCA1 in a lipid membrane enabled us to gain important mechanistic and structural insights and paves the way for investigating modulators that target the functions of ABCA1. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27079.map.gz emd_27079.map.gz | 58.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27079-v30.xml emd-27079-v30.xml emd-27079.xml emd-27079.xml | 19.1 KB 19.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_27079_fsc.xml emd_27079_fsc.xml | 11.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_27079.png emd_27079.png | 23.3 KB | ||
| Filedesc metadata |  emd-27079.cif.gz emd-27079.cif.gz | 6.4 KB | ||
| Others |  emd_27079_half_map_1.map.gz emd_27079_half_map_1.map.gz emd_27079_half_map_2.map.gz emd_27079_half_map_2.map.gz | 49.7 MB 49.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27079 http://ftp.pdbj.org/pub/emdb/structures/EMD-27079 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27079 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27079 | HTTPS FTP |
-Validation report
| Summary document |  emd_27079_validation.pdf.gz emd_27079_validation.pdf.gz | 844 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_27079_full_validation.pdf.gz emd_27079_full_validation.pdf.gz | 843.5 KB | Display | |
| Data in XML |  emd_27079_validation.xml.gz emd_27079_validation.xml.gz | 16.1 KB | Display | |
| Data in CIF |  emd_27079_validation.cif.gz emd_27079_validation.cif.gz | 21.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27079 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27079 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27079 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27079 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_27079.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27079.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.24 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_27079_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_27079_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Human ABCA1 reconstituted into nanodiscs
| Entire | Name: Human ABCA1 reconstituted into nanodiscs |
|---|---|
| Components |
|
-Supramolecule #1: Human ABCA1 reconstituted into nanodiscs
| Supramolecule | Name: Human ABCA1 reconstituted into nanodiscs / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: ABCA1 in nanodisc
| Macromolecule | Name: ABCA1 in nanodisc / type: protein_or_peptide / ID: 1 / Enantiomer: DEXTRO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GGGASEFVDM ACWPQLRLLL WKNLTFRRRQ TCQLLLEVAW PLFIFLILIS VRLSYPPYEQ HECHFPNKA MPSAGTLPWV QGIICNANNP CFRYPTPGEA PGVVGNFNKS IVARLFSDAR RLLLYSQKDT SMKDMRKVL RTLQQIKKSS SNLKLQDFLV DNETFSGFLY ...String: GGGASEFVDM ACWPQLRLLL WKNLTFRRRQ TCQLLLEVAW PLFIFLILIS VRLSYPPYEQ HECHFPNKA MPSAGTLPWV QGIICNANNP CFRYPTPGEA PGVVGNFNKS IVARLFSDAR RLLLYSQKDT SMKDMRKVL RTLQQIKKSS SNLKLQDFLV DNETFSGFLY HNLSLPKSTV DKMLRADVIL H KVFLQGYQ LHLTSLCNGS KSEEMIQLGD QEVSELCGLP REKLAAAERV LRSNMDILKP IL RTLNSTS PFPSKELAEA TKTLLHSLGT LAQELFSMRS WSDMRQEVMF LTNVNSSSSS TQI YQAVSR IVCGHPEGGG LKIKSLNWYE DNNYKALFGG NGTEEDAETF YDNSTTPYCN DLMK NLESS PLSRIIWKAL KPLLVGKILY TPDTPATRQV MAEVNKTFQE LAVFHDLEGM WEELS PKIW TFMENSQEMD LVRMLLDSRD NDHFWEQQLD GLDWTAQDIV AFLAKHPEDV QSSNGS VYT WREAFNETNQ AIRTISRFME CVNLNKLEPI ATEVWLINKS MELLDERKFW AGIVFTG IT PGSIELPHHV KYKIRMDIDN VERTNKIKDG YWDPGPRADP FEDMRYVWGG FAYLQDVV E QAIIRVLTGT EKKTGVYMQQ MPYPCYVDDI FLRVMSRSMP LFMTLAWIYS VAVIIKGIV YEKEARLKET MRIMGLDNSI LWFSWFISSL IPLLVSAGLL VVILKLGNLL PYSDPSVVFV FLSVFAVVT ILQCFLISTL FSRANLAAAC GGIIYFTLYL PYVLCVAWQD YVGFTLKIFA S LLSPVAFG FGCEYFALFE EQGIGVQWDN LFESPVEEDG FNLTTSVSMM LFDTFLYGVM TW YIEAVFP GQYGIPRPWY FPCTKSYWFG EESDEKSHPG SNQKRISEIC MEEEPTHLKL GVS IQNLVK VYRDGMKVAV DGLALNFYEG QITSFLGHNG AGKTTTMSIL TGLFPPTSGT AYIL GKDIR SEMSTIRQNL GVCPQHNVLF DMLTVEEHIW FYARLKGLSE KHVKAEMEQM ALDVG LPSS KLKSKTSQLS GGMQRKLSVA LAFVGGSKVV ILDEPTAGVD PYSRRGIWEL LLKYRQ GRT IILSTHHMDE ADVLGDRIAI ISHGKLCCVG SSLFLKNQLG TGYYLTLVKK DVESSLS SC RNSSSTVSYL KKEDSVSQSS SDAGLGSDHE SDTLTIDVSA ISNLIRKHVS EARLVEDI G HELTYVLPYE AAKEGAFVEL FHEIDDRLSD LGISSYGISE TTLEEIFLKV AEESGVDAE TSDGTLPARR NRRAFGDKQS CLRPFTEDDA ADPNDSDIDP ESRETDLLSG MDGKGSYQVK GWKLTQQQF VALLWKRLLI ARRSRKGFFA QIVLPAVFVC IALVFSLIVP PFGKYPSLEL Q PWMYNEQY TFVSNDAPED TGTLELLNAL TKDPGFGTRC MEGNPIPDTP CQAGEEEWTT AP VPQTIMD LFQNGNWTMQ NPSPACQCSS DKIKKMLPVC PPGAGGLPPP QRKQNTADIL QDL TGRNIS DYLVKTYVQI IAKSLKNKIW VNEFRYGGFS LGVSNTQALP PSQEVNDAIK QMKK HLKLA KDSSADRFLN SLGRFMTGLD TKNNVKVWFN NKGWHAISSF LNVINNAILR ANLQK GENP SHYGITAFNH PLNLTKQQLS EVALMTTSVD VLVSICVIFA MSFVPASFVV FLIQER VSK AKHLQFISGV KPVIYWLSNF VWDMCNYVVP ATLVIIIFIC FQQKSYVSST NLPVLAL LL LLYGWSITPL MYPASFVFKI PSTAYVVLTS VNLFIGINGS VATFVLELFT DNKLNNIN D ILKSVFLIFP HFCLGRGLID MVKNQAMADA LERFGENRFV SPLSWDLVGR NLFAMAVEG VVFFLITVLI QYRFFIRPRP VNAKLSPLND EDEDVRRERQ RILDGGGQND ILEIKELTKI YRRKRKPAV DRICVGIPPG ECFGLLGVNG AGKSSTFKML TGDTTVTRGD AFLNKNSILS N IHEVHQNM GYCPQFDAIT ELLTGREHVE FFALLRGVPE KEVGKVGEWA IRKLGLVKYG EK YAGNYSG GNKRKLSTAM ALIGGPPVVF LDEPTTGMDP KARRFLWNCA LSVVKEGRSV VLT SHSMEE CEALCTRMAI MVNGRFRCLG SVQHLKNRFG DGYTIVVRIA GSNPDLKPVQ DFFG LAFPG SVLKEKHRNM LQYQLPSSLS SLARIFSILS QSKKRLHIED YSVSQTTLDQ VFVNF AKDQ SDDDHLKDLS LHKNQTVVDV AVLTSFLQDE KVKESYV |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.8 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||||||
| Vitrification | Cryogen name: ETHANE | ||||||||||||
| Details | Homogeneous, monodisperse sample of ABCA1 in nanodiscs |
- Electron microscopy #1
Electron microscopy #1
| Microscopy ID | 1 |
|---|---|
| Microscope | FEI TITAN KRIOS |
| Image recording | Image recording ID: 1 / Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 1.04 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Electron microscopy #1~
Electron microscopy #1~
| Microscopy ID | 1 |
|---|---|
| Microscope | FEI TALOS ARCTICA |
| Image recording | Image recording ID: 2 / Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 1.07 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 36000 |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)