[English] 日本語
 Yorodumi
Yorodumi- EMDB-26981: Cryo-EM structure of Mtb Lpd bound to inhibitor complex with 2-((... -
+ Open data
Open data
- Basic information
Basic information
| Entry | 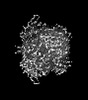 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Mtb Lpd bound to inhibitor complex with 2-((2-cyano-N,5-dimethyl-1H-indole)-7-sulfonamido)-N-(4-(oxetan-3-yl)-3,4-dihydro-2H-benzo[b] [1,4]oxazin-7-yl)acetamide | ||||||||||||
 Map data Map data | Density-modified map generated using phenix.cryoem.resolve | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | flavoprotein / glycolysis / redox-active center / OXIDOREDUCTASE-INHIBITOR complex | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationCell redox homeostasis / dihydrolipoyl dehydrogenase / dihydrolipoyl dehydrogenase (NADH) activity / NADH binding / disulfide oxidoreductase activity / pyruvate dehydrogenase complex / 2-oxoglutarate metabolic process / pyruvate metabolic process / zymogen binding / oxidoreductase activity, acting on NAD(P)H, quinone or similar compound as acceptor ...Cell redox homeostasis / dihydrolipoyl dehydrogenase / dihydrolipoyl dehydrogenase (NADH) activity / NADH binding / disulfide oxidoreductase activity / pyruvate dehydrogenase complex / 2-oxoglutarate metabolic process / pyruvate metabolic process / zymogen binding / oxidoreductase activity, acting on NAD(P)H, quinone or similar compound as acceptor / antioxidant activity / Prevention of phagosomal-lysosomal fusion / cell redox homeostasis / flavin adenine dinucleotide binding / extracellular region / plasma membrane / cytosol Similarity search - Function | ||||||||||||
| Biological species |  | ||||||||||||
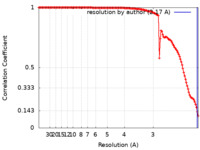
| Method | single particle reconstruction / cryo EM / Resolution: 2.17 Å | ||||||||||||
 Authors Authors | Kochanczyk T / Arango N / Lima CD | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Not published Journal: Not publishedTitle: Cryo-EM structure of Mtb Lpd bound to the inhibitor 2-((2-cyano-N,5-dimethyl-1H-indole)-7-sulfonamido)-N-(4-(oxetan-3-yl)-3,4-dihydro-2H-benzo[b] [1,4]oxazin-7-yl)acetamide at 2.17 Angstrom resolution Authors: Kochanczyk T / Arango N / Michino M / Sun S / Ginn J / Bryk R / Nathan C / Lima CD | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26981.map.gz emd_26981.map.gz | 3.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26981-v30.xml emd-26981-v30.xml emd-26981.xml emd-26981.xml | 21 KB 21 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_26981_fsc.xml emd_26981_fsc.xml | 13.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_26981.png emd_26981.png | 76.3 KB | ||
| Filedesc metadata |  emd-26981.cif.gz emd-26981.cif.gz | 6.5 KB | ||
| Others |  emd_26981_additional_1.map.gz emd_26981_additional_1.map.gz emd_26981_half_map_1.map.gz emd_26981_half_map_1.map.gz emd_26981_half_map_2.map.gz emd_26981_half_map_2.map.gz | 109.6 MB 200.3 MB 200.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26981 http://ftp.pdbj.org/pub/emdb/structures/EMD-26981 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26981 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26981 | HTTPS FTP |
-Related structure data
| Related structure data |  8ct4MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_26981.map.gz / Format: CCP4 / Size: 4.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26981.map.gz / Format: CCP4 / Size: 4.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Density-modified map generated using phenix.cryoem.resolve | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.064 Å | ||||||||||||||||||||||||||||||||||||
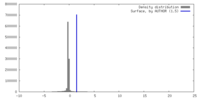
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Raw map
| File | emd_26981_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Raw map | ||||||||||||
| Projections & Slices |
| ||||||||||||
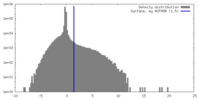
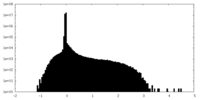
| Density Histograms |
-Half map: half-map 1
| File | emd_26981_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half-map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
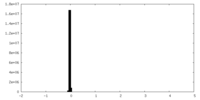
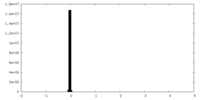
| Density Histograms |
-Half map: half-map 2
| File | emd_26981_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half-map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
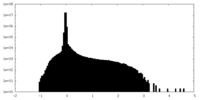
| Density Histograms |
- Sample components
Sample components
-Entire : Dihydrolipoyl dehydrogenase in complex with 2-((2-cyano-N,5-dimet...
| Entire | Name: Dihydrolipoyl dehydrogenase in complex with 2-((2-cyano-N,5-dimethyl-1H-indole)-7-sulfonamido)-N-(4-(oxetan-3-yl)-3,4-dihydro-2H-benzo[b] [1,4]oxazin-7-yl)acetamide |
|---|---|
| Components |
|
-Supramolecule #1: Dihydrolipoyl dehydrogenase in complex with 2-((2-cyano-N,5-dimet...
| Supramolecule | Name: Dihydrolipoyl dehydrogenase in complex with 2-((2-cyano-N,5-dimethyl-1H-indole)-7-sulfonamido)-N-(4-(oxetan-3-yl)-3,4-dihydro-2H-benzo[b] [1,4]oxazin-7-yl)acetamide type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 Details: 2-((2-cyano-N,5-dimethyl-1H-indole)-7-sulfonamido)-N-(4-(oxetan-3-yl)-3,4-dihydro-2H-benzo[b] [1,4]oxazin-7-yl)acetamide provided by the Tri-Institutional Therapeutics Discovery Institute ...Details: 2-((2-cyano-N,5-dimethyl-1H-indole)-7-sulfonamido)-N-(4-(oxetan-3-yl)-3,4-dihydro-2H-benzo[b] [1,4]oxazin-7-yl)acetamide provided by the Tri-Institutional Therapeutics Discovery Institute (tritdi.org) contact: John Ginn (jginn@tritdi.org) |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 99 KDa |
-Macromolecule #1: Dihydrolipoyl dehydrogenase
| Macromolecule | Name: Dihydrolipoyl dehydrogenase / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: dihydrolipoyl dehydrogenase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 49.437035 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GSMTHYDVVV LGAGPGGYVA AIRAAQLGLS TAIVEPKYWG GVCLNVGCIP SKALLRNAEL VHIFTKDAKA FGISGEVTFD YGIAYDRSR KVAEGRVAGV HFLMKKNKIT EIHGYGTFAD ANTLLVDLND GGTESVTFDN AIIATGSSTR LVPGTSLSAN V VTYEEQIL ...String: GSMTHYDVVV LGAGPGGYVA AIRAAQLGLS TAIVEPKYWG GVCLNVGCIP SKALLRNAEL VHIFTKDAKA FGISGEVTFD YGIAYDRSR KVAEGRVAGV HFLMKKNKIT EIHGYGTFAD ANTLLVDLND GGTESVTFDN AIIATGSSTR LVPGTSLSAN V VTYEEQIL SRELPKSIII AGAGAIGMEF GYVLKNYGVD VTIVEFLPRA LPNEDADVSK EIEKQFKKLG VTILTATKVE SI ADGGSQV TVTVTKDGVA QELKAEKVLQ AIGFAPNVEG YGLDKAGVAL TDRKAIGVDD YMRTNVGHIY AIGDVNGLLQ LAH VAEAQG VVAAETIAGA ETLTLGDHRM LPRATFCQPN VASFGLTEQQ ARNEGYDVVV AKFPFTANAK AHGVGDPSGF VKLV ADAKH GELLGGHLVG HDVAELLPEL TLAQRWDLTA SELARNVHTH PTMSEALQEC FHGLVGHMIN F UniProtKB: Dihydrolipoyl dehydrogenase |
-Macromolecule #2: FLAVIN-ADENINE DINUCLEOTIDE
| Macromolecule | Name: FLAVIN-ADENINE DINUCLEOTIDE / type: ligand / ID: 2 / Number of copies: 2 / Formula: FAD |
|---|---|
| Molecular weight | Theoretical: 785.55 Da |
| Chemical component information |  ChemComp-FAD: |
-Macromolecule #3: N~2~-(2-cyano-5-methyl-1H-indole-7-sulfonyl)-N~2~-methyl-N-[4-(ox...
| Macromolecule | Name: N~2~-(2-cyano-5-methyl-1H-indole-7-sulfonyl)-N~2~-methyl-N-[4-(oxetan-3-yl)-3,4-dihydro-2H-1,4-benzoxazin-7-yl]glycinamide type: ligand / ID: 3 / Number of copies: 2 / Formula: OU6 |
|---|---|
| Molecular weight | Theoretical: 495.551 Da |
| Chemical component information | 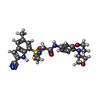 ChemComp-OU6: |
-Macromolecule #4: water
| Macromolecule | Name: water / type: ligand / ID: 4 / Number of copies: 665 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3 mg/mL |
|---|---|
| Buffer | pH: 8 / Details: 20 mM Tris-HCl, pH 8.0, 87.5 mM NaCl, 0.05% IGEPAL |
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV Details: Temperature 22 C, humidity 100%, Wait time 8s, Blot time: 3.5s. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Detector mode: SUPER-RESOLUTION / Number grids imaged: 1 / Number real images: 10748 / Average exposure time: 4.0 sec. / Average electron dose: 70.66 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 22500 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Chain ID: AB / Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Software | Name: PHENIX (ver. 1.20.1-44487) |
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Target criteria: Correlation coefficient |
| Output model |  PDB-8ct4: |
 Movie
Movie Controller
Controller








 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)