[English] 日本語
 Yorodumi
Yorodumi- EMDB-26848: Geometrically programmed DNA origami colloid for self-assembly of... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Geometrically programmed DNA origami colloid for self-assembly of tubules: X-particle | |||||||||
 Map data Map data | DNA origami particle for tubule self-assembly | |||||||||
 Sample Sample |
| |||||||||
| Biological species | synthetic construct (others) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 23.5 Å | |||||||||
 Authors Authors | Hayakawa D / Videbaek TE / Hall DM / Fang H / Sigl C / Feigl E / Dietz H / Fraden S / Hagan MF / Grason GM / Rogers WB | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2022 Journal: Proc Natl Acad Sci U S A / Year: 2022Title: Geometrically programmed self-limited assembly of tubules using DNA origami colloids. Authors: Daichi Hayakawa / Thomas E Videbaek / Douglas M Hall / Huang Fang / Christian Sigl / Elija Feigl / Hendrik Dietz / Seth Fraden / Michael F Hagan / Gregory M Grason / W Benjamin Rogers /   Abstract: Self-assembly is one of the most promising strategies for making functional materials at the nanoscale, yet new design principles for making self-limiting architectures, rather than spatially ...Self-assembly is one of the most promising strategies for making functional materials at the nanoscale, yet new design principles for making self-limiting architectures, rather than spatially unlimited periodic lattice structures, are needed. To address this challenge, we explore the tradeoffs between addressable assembly and self-closing assembly of a specific class of self-limiting structures: cylindrical tubules. We make triangular subunits using DNA origami that have specific, valence-limited interactions and designed binding angles, and we study their assembly into tubules that have a self-limited width that is much larger than the size of an individual subunit. In the simplest case, the tubules are assembled from a single component by geometrically programming the dihedral angles between neighboring subunits. We show that the tubules can reach many micrometers in length and that their average width can be prescribed through the dihedral angles. We find that there is a distribution in the width and the chirality of the tubules, which we rationalize by developing a model that considers the finite bending rigidity of the assembled structure as well as the mechanism of self-closure. Finally, we demonstrate that the distributions of tubules can be further sculpted by increasing the number of subunit species, thereby increasing the assembly complexity, and demonstrate that using two subunit species successfully reduces the number of available end states by half. These results help to shed light on the roles of assembly complexity and geometry in self-limited assembly and could be extended to other self-limiting architectures, such as shells, toroids, or triply periodic frameworks. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26848.map.gz emd_26848.map.gz | 166.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26848-v30.xml emd-26848-v30.xml emd-26848.xml emd-26848.xml | 15.6 KB 15.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_26848_fsc.xml emd_26848_fsc.xml | 13 KB | Display |  FSC data file FSC data file |
| Images |  emd_26848.png emd_26848.png | 81.2 KB | ||
| Others |  emd_26848_half_map_1.map.gz emd_26848_half_map_1.map.gz emd_26848_half_map_2.map.gz emd_26848_half_map_2.map.gz | 140.7 MB 141 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26848 http://ftp.pdbj.org/pub/emdb/structures/EMD-26848 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26848 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26848 | HTTPS FTP |
-Validation report
| Summary document |  emd_26848_validation.pdf.gz emd_26848_validation.pdf.gz | 806 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_26848_full_validation.pdf.gz emd_26848_full_validation.pdf.gz | 805.6 KB | Display | |
| Data in XML |  emd_26848_validation.xml.gz emd_26848_validation.xml.gz | 20 KB | Display | |
| Data in CIF |  emd_26848_validation.cif.gz emd_26848_validation.cif.gz | 26.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26848 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26848 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26848 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26848 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_26848.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26848.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | DNA origami particle for tubule self-assembly | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.87 Å | ||||||||||||||||||||||||||||||||||||
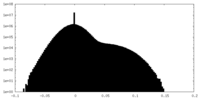
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half map (1) from Refine 3D (RELION)
| File | emd_26848_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map (1) from Refine 3D (RELION) | ||||||||||||
| Projections & Slices |
| ||||||||||||
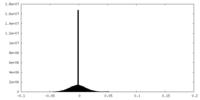
| Density Histograms |
-Half map: Half map (2) from Refine 3D (RELION)
| File | emd_26848_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map (2) from Refine 3D (RELION) | ||||||||||||
| Projections & Slices |
| ||||||||||||
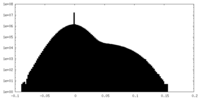
| Density Histograms |
- Sample components
Sample components
-Entire : Geometrically programmed DNA origami colloid for self-assembly of...
| Entire | Name: Geometrically programmed DNA origami colloid for self-assembly of tubules: X-particle |
|---|---|
| Components |
|
-Supramolecule #1: Geometrically programmed DNA origami colloid for self-assembly of...
| Supramolecule | Name: Geometrically programmed DNA origami colloid for self-assembly of tubules: X-particle type: complex / Chimera: Yes / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Grid | Model: C-flat-1.2/1.3 / Material: COPPER / Mesh: 400 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 293 K / Instrument: FEI VITROBOT MARK I |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F30 |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Detector mode: INTEGRATING / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Average exposure time: 4.0 sec. / Average electron dose: 31.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 150.0 µm / Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 2.0 µm / Nominal magnification: 39000 |
| Sample stage | Specimen holder model: GATAN 626 SINGLE TILT LIQUID NITROGEN CRYO TRANSFER HOLDER Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Tecnai F30 / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)