+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | KSQ+AT from first module of the pikromycin synthase | |||||||||
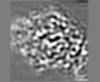
 Map data Map data | Primary map. Density fades for distal AT domains. Use AT_newest.ccp4 for these (related through twofold of PikKSAT1_dimer.cif). | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ketosynthase-like / acyltransferase / decarboxylation / BIOSYNTHETIC PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology information10-deoxymethynolide synthase / narbonolide synthase / macrolide biosynthetic process / acyltransferase activity, transferring groups other than amino-acyl groups / fatty acid synthase activity / phosphopantetheine binding / 3-oxoacyl-[acyl-carrier-protein] synthase activity / fatty acid biosynthetic process Similarity search - Function | |||||||||
| Biological species |  Streptomyces venezuelae (bacteria) Streptomyces venezuelae (bacteria) | |||||||||
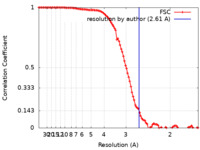
| Method | single particle reconstruction / cryo EM / Resolution: 2.61 Å | |||||||||
 Authors Authors | Keatinge-Clay AT / Dickinson MS / Miyazawa T / McCool RS | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Structure / Year: 2022 Journal: Structure / Year: 2022Title: Priming enzymes from the pikromycin synthase reveal how assembly-line ketosynthases catalyze carbon-carbon chemistry. Authors: Miles S Dickinson / Takeshi Miyazawa / Ryan S McCool / Adrian T Keatinge-Clay /  Abstract: The first domain of modular polyketide synthases (PKSs) is most commonly a ketosynthase (KS)-like enzyme, KS, that primes polyketide synthesis. Unlike downstream KSs that fuse α-carboxyacyl groups ...The first domain of modular polyketide synthases (PKSs) is most commonly a ketosynthase (KS)-like enzyme, KS, that primes polyketide synthesis. Unlike downstream KSs that fuse α-carboxyacyl groups to growing polyketide chains, it performs an extension-decoupled decarboxylation of these groups to generate primer units. When Pik127, a model triketide synthase constructed from modules of the pikromycin synthase, was studied by cryoelectron microscopy (cryo-EM), the dimeric didomain comprised of KS and the neighboring methylmalonyl-selective acyltransferase (AT) dominated the class averages and yielded structures at 2.5- and 2.8-Å resolution, respectively. Comparisons with ketosynthases complexed with their substrates revealed the conformation of the (2S)-methylmalonyl-S-phosphopantetheinyl portion of KS and KS substrates prior to decarboxylation. Point mutants of Pik127 probed the roles of residues in the KS active site, while an AT-swapped version of Pik127 demonstrated that KS can also decarboxylate malonyl groups. Mechanisms for how KS and KS domains catalyze carbon-carbon chemistry are proposed. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26839.map.gz emd_26839.map.gz | 4.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26839-v30.xml emd-26839-v30.xml emd-26839.xml emd-26839.xml | 23.1 KB 23.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_26839_fsc.xml emd_26839_fsc.xml | 10.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_26839.png emd_26839.png | 67.7 KB | ||
| Filedesc metadata |  emd-26839.cif.gz emd-26839.cif.gz | 7.3 KB | ||
| Others |  emd_26839_additional_1.map.gz emd_26839_additional_1.map.gz emd_26839_half_map_1.map.gz emd_26839_half_map_1.map.gz emd_26839_half_map_2.map.gz emd_26839_half_map_2.map.gz | 1 MB 95.3 MB 95.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26839 http://ftp.pdbj.org/pub/emdb/structures/EMD-26839 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26839 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26839 | HTTPS FTP |
-Related structure data
| Related structure data |  7uwrMC  8czcC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_26839.map.gz / Format: CCP4 / Size: 5.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26839.map.gz / Format: CCP4 / Size: 5.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Primary map. Density fades for distal AT domains. Use AT_newest.ccp4 for these (related through twofold of PikKSAT1_dimer.cif). | ||||||||||||||||||||||||||||||||||||
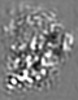
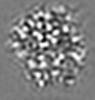
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.81 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Map for AT (Chain B) obtained through local...
| File | emd_26839_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map for AT (Chain B) obtained through local refinement. Apply twofold from PikKSQAT1.cif to obtain map for AT (Chain A). | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: second half map
| File | emd_26839_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | second half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: first half map
| File | emd_26839_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | first half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Pik127
| Entire | Name: Pik127 |
|---|---|
| Components |
|
-Supramolecule #1: Pik127
| Supramolecule | Name: Pik127 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 Details: Polyketide synthase engineered from the first, second, and seventh modules of the pikromycin synthase as described in Miyazawa et al., 2021, Chem. Commun. 57:8762-8765 |
|---|---|
| Source (natural) | Organism:  Streptomyces venezuelae (bacteria) / Strain: 15439 Streptomyces venezuelae (bacteria) / Strain: 15439 |
| Molecular weight | Theoretical: 410 KDa |
-Macromolecule #1: Narbonolide/10-deoxymethynolide synthase PikA1, modules 1 and 2
| Macromolecule | Name: Narbonolide/10-deoxymethynolide synthase PikA1, modules 1 and 2 type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: 10-deoxymethynolide synthase |
|---|---|
| Source (natural) | Organism:  Streptomyces venezuelae (bacteria) Streptomyces venezuelae (bacteria) |
| Molecular weight | Theoretical: 91.797898 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: EPVAVVGISC RVPGARDPRE FWELLAAGGQ AVTDVPADRW NAGDFYDPDR SAPGRSNSRW GGFIEDVDRF DAAFFGISPR EAAEMDPQQ RLALELGWEA LERAGIDPSS LTGTRTGVFA GAIWDDYATL KHRQGGAAIT PHTVTGLHRG IIANRLSYTL G LRGPSMVV ...String: EPVAVVGISC RVPGARDPRE FWELLAAGGQ AVTDVPADRW NAGDFYDPDR SAPGRSNSRW GGFIEDVDRF DAAFFGISPR EAAEMDPQQ RLALELGWEA LERAGIDPSS LTGTRTGVFA GAIWDDYATL KHRQGGAAIT PHTVTGLHRG IIANRLSYTL G LRGPSMVV DSGQSSSLVA VHLACESLRR GESELALAGG VSLNLVPDSI IGASKFGGLS PDGRAYTFDA RANGYVRGEG GG FVVLKRL SRAVADGDPV LAVIRGSAVN NGGAAQGMTT PDAQAQEAVL REAHERAGTA PADVRYVELH GTGTPVGDPI EAA ALGAAL GTGRPAGQPL LVGSVKTNIG HLEGAAGIAG LIKAVLAVRG RALPASLNYE TPNPAIPFEE LNLRVNTEYL PWEP EHDGQ RMVVGVSSFG MGGTNAHVVL EEAPGVVEGA SVVESTVGGS AVGGGVVPWV VSAKSAAALD AQIERLAAFA SRDRT DGVD AGAVDAGAVD AGAVARVLAG GRAQFEHRAV VVGSGPDDLA AALAAPEGLV RGVASGVGRV AFVFPGQGTQ WAGMGA ELL DSSAVFAAAM AECEAALSPY VDWSLEAVVR QAPGAPTLER VDVVQPVTFA VMVSLARVWQ HHGVTPQAVV GHSQGEI AA AYVAGALSLD DAARVVTLRS KSIAAHLAGK GGMLSLALSE DAVLERLAGF DGLSVAAVNG PTATVVSGDP VQIEELAR A CEADGVRARV IPVDYASHSR QVEIIESELA EVLAGLSPQA PRVPFFSTLE GAWITEPVLD GGYWYRNLRH RVGFAPAVE TLATDEGFTH FVEVSAHPVL TMALPGTVTG LATLRRDNGG QDRLVASLAE AWANGLAVDW SPLLPSATGH HSDLPTYAFQ TERHWL UniProtKB: Pikromycin polyketide synthase component PikAI |
-Macromolecule #2: water
| Macromolecule | Name: water / type: ligand / ID: 2 / Number of copies: 55 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.5 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
Details: Buffer was made fresh because TCEP rapidly degrades in phosphate | |||||||||||||||
| Grid | Model: C-flat-2/2 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Support film - Film thickness: 12 / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 30 sec. | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV Details: Blotted for 3 seconds with a force of "1" before plunging.. | |||||||||||||||
| Details | This sample was monodisperse. It was incubated with its substrates, methylmalonyl-CoA and NADPH, for 30 min. at 25 C before freezing. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Details | Collected 4284 movies at 0 degrees tilt and 2912 movies at -30 degrees tilt |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 7196 / Average exposure time: 5.0 sec. / Average electron dose: 70.0 e/Å2 / Details: 20 frames per second |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated magnification: 61728 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.8 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)