[English] 日本語
 Yorodumi
Yorodumi- EMDB-26659: Prefusion-stabilized Nipah virus fusion protein complexed with Fab 1H8 -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Prefusion-stabilized Nipah virus fusion protein complexed with Fab 1H8 | |||||||||
 Map data Map data | Prefusion-stabilized Nipah virus fusion protein complexed with Fab 1H8 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Henipavirus / Nipah virus / NiV / F / fusion / prefusion / preF / pre-F / neutralizing antibody / Fab / VIRAL PROTEIN / VIRAL PROTEIN-Immune System complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationmembrane fusion involved in viral entry into host cell / fusion of virus membrane with host plasma membrane / viral envelope / symbiont entry into host cell / host cell plasma membrane / virion membrane / membrane Similarity search - Function | |||||||||
| Biological species |  Nipah henipavirus / Nipah henipavirus /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.5 Å | |||||||||
 Authors Authors | Byrne PO / McLellan JS | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Structural basis for antibody recognition of vulnerable epitopes on Nipah virus F protein. Authors: Patrick O Byrne / Brian E Fisher / David R Ambrozak / Elizabeth G Blade / Yaroslav Tsybovsky / Barney S Graham / Jason S McLellan / Rebecca J Loomis /   Abstract: Nipah virus (NiV) is a pathogenic paramyxovirus that causes fatal encephalitis in humans. Two envelope glycoproteins, the attachment protein (G/RBP) and fusion protein (F), facilitate entry into host ...Nipah virus (NiV) is a pathogenic paramyxovirus that causes fatal encephalitis in humans. Two envelope glycoproteins, the attachment protein (G/RBP) and fusion protein (F), facilitate entry into host cells. Due to its vital role, NiV F presents an attractive target for developing vaccines and therapeutics. Several neutralization-sensitive epitopes on the NiV F apex have been described, however the antigenicity of most of the F protein's surface remains uncharacterized. Here, we immunize mice with prefusion-stabilized NiV F and isolate ten monoclonal antibodies that neutralize pseudotyped virus. Cryo-electron microscopy reveals eight neutralization-sensitive epitopes on NiV F, four of which have not previously been described. Novel sites span the lateral and basal faces of NiV F, expanding the known library of vulnerable epitopes. Seven of ten antibodies bind the Hendra virus (HeV) F protein. Multiple sequence alignment suggests that some of these newly identified neutralizing antibodies may also bind F proteins across the Henipavirus genus. This work identifies new epitopes as targets for therapeutics, provides a molecular basis for NiV neutralization, and lays a foundation for development of new cross-reactive antibodies targeting Henipavirus F proteins. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26659.map.gz emd_26659.map.gz | 290.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26659-v30.xml emd-26659-v30.xml emd-26659.xml emd-26659.xml | 22.1 KB 22.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_26659_fsc.xml emd_26659_fsc.xml | 16.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_26659.png emd_26659.png | 58.7 KB | ||
| Masks |  emd_26659_msk_1.map emd_26659_msk_1.map | 307.5 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-26659.cif.gz emd-26659.cif.gz | 6.7 KB | ||
| Others |  emd_26659_additional_1.map.gz emd_26659_additional_1.map.gz emd_26659_half_map_1.map.gz emd_26659_half_map_1.map.gz emd_26659_half_map_2.map.gz emd_26659_half_map_2.map.gz | 154.2 MB 285.6 MB 285.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26659 http://ftp.pdbj.org/pub/emdb/structures/EMD-26659 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26659 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26659 | HTTPS FTP |
-Validation report
| Summary document |  emd_26659_validation.pdf.gz emd_26659_validation.pdf.gz | 789.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_26659_full_validation.pdf.gz emd_26659_full_validation.pdf.gz | 789.3 KB | Display | |
| Data in XML |  emd_26659_validation.xml.gz emd_26659_validation.xml.gz | 23.3 KB | Display | |
| Data in CIF |  emd_26659_validation.cif.gz emd_26659_validation.cif.gz | 30.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26659 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26659 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26659 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26659 | HTTPS FTP |
-Related structure data
| Related structure data |  7upaMC  7uopC  7up9C  7upbC  7updC  7upkC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_26659.map.gz / Format: CCP4 / Size: 307.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26659.map.gz / Format: CCP4 / Size: 307.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Prefusion-stabilized Nipah virus fusion protein complexed with Fab 1H8 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_26659_msk_1.map emd_26659_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
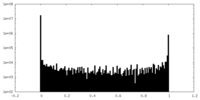
| Density Histograms |
-Additional map: Additional map 1
| File | emd_26659_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Additional map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
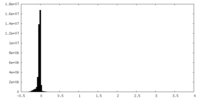
| Density Histograms |
-Half map: Half Map 1
| File | emd_26659_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
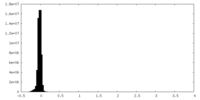
| Density Histograms |
-Half map: Half Map 2
| File | emd_26659_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
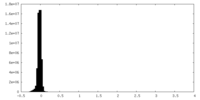
| Density Histograms |
- Sample components
Sample components
-Entire : Prefusion-stabilized Nipah virus fusion protein complexed with Fab 1H8
| Entire | Name: Prefusion-stabilized Nipah virus fusion protein complexed with Fab 1H8 |
|---|---|
| Components |
|
-Supramolecule #1: Prefusion-stabilized Nipah virus fusion protein complexed with Fab 1H8
| Supramolecule | Name: Prefusion-stabilized Nipah virus fusion protein complexed with Fab 1H8 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Nipah henipavirus Nipah henipavirus |
-Macromolecule #1: Fab 1H8 heavy chain
| Macromolecule | Name: Fab 1H8 heavy chain / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 15.335375 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MEFGLSWIFL AAILKGVQCE VQLQQSGPEL VKPGASVKIS CKASGYSFTG YTMNWVKQSH GKNLEWIGLI NPFIGGTRYN QKFKGKATL TVDKSSRTAY MELLSLTSED SAVYYCAREA DYDWYFDVWG AGTTVTVSS |
-Macromolecule #2: Fab 1H8 light chain
| Macromolecule | Name: Fab 1H8 light chain / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 13.880542 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MEFGLSWIFL AAILKGVQCD IQMTQTTSSL SASLGDRVTI SCRASQDISN YLHWYQQKPD GTVNLLIFYT SRLHSGVPSR FSGSGSGTD YSLTISNLEQ EDIATYFCQQ GNTLPRTFGG GTKLEIK |
-Macromolecule #3: Fusion glycoprotein F0
| Macromolecule | Name: Fusion glycoprotein F0 / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Nipah henipavirus Nipah henipavirus |
| Molecular weight | Theoretical: 52.893848 KDa |
| Recombinant expression | Organism:  Nipah henipavirus Nipah henipavirus |
| Sequence | String: MVVILDKRCY CNLLILILMI SECSVGILHY EKLSKIGLVK GVTRKYKIKS NPLTKDIVIK MIPNVSNMSQ CTGSVMENYK TRLNGILTP IKGALEIYKN NTHDCVGDVR LAGVCMAGVA IGIATAAQIT AGVALYEAMK NADNINKLKS SIESTNEAVV K LQETAEKT ...String: MVVILDKRCY CNLLILILMI SECSVGILHY EKLSKIGLVK GVTRKYKIKS NPLTKDIVIK MIPNVSNMSQ CTGSVMENYK TRLNGILTP IKGALEIYKN NTHDCVGDVR LAGVCMAGVA IGIATAAQIT AGVALYEAMK NADNINKLKS SIESTNEAVV K LQETAEKT VYVFTALQDY INTNLVPTID KIPCKQTELS LDLALSKYLS DLLFVFGPNL QDPVSNSMTI QAISQAFGGN YE TLLRTLG YATEDFDDLL ESDSITGQII YVDLSSYYII VRVYFPILTE IQQAYIQELL PVSFNNDNSE WISIVPNFIL VRN TLISNI EIGFCLITKR SVICNQDYAT PMTNNMRECL TGSTEKCPRE LVVSSHVPRF ALSNGVLFAN CISVTCQCQT TGRA ISQSG EQTLLMIDNT TCPTAVLGNV IISLGKYLGS VNYNSEGIAI GPPVFTDKVD ISSQISSMNQ SLQQSKDYIK EAQRL UniProtKB: Fusion glycoprotein F0 |
-Macromolecule #4: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 4 / Number of copies: 12 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 80.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)