+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human TMEM175 in an closed state | ||||||||||||
 Map data Map data | Closed TMEM175 | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | potassium channel / MEMBRANE PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationlysosomal lumen pH elevation / phagosome-lysosome fusion / regulation of lysosomal lumen pH / potassium ion leak channel activity / proton channel activity / arachidonate binding / potassium channel activity / potassium ion transmembrane transport / proton transmembrane transport / neuron cellular homeostasis ...lysosomal lumen pH elevation / phagosome-lysosome fusion / regulation of lysosomal lumen pH / potassium ion leak channel activity / proton channel activity / arachidonate binding / potassium channel activity / potassium ion transmembrane transport / proton transmembrane transport / neuron cellular homeostasis / lysosome / endosome / endosome membrane / lysosomal membrane Similarity search - Function | ||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.61 Å | ||||||||||||
 Authors Authors | Oh S / Hite RK | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Elife / Year: 2022 Journal: Elife / Year: 2022Title: Differential ion dehydration energetics explains selectivity in the non-canonical lysosomal K channel TMEM175. Authors: SeCheol Oh / Fabrizio Marinelli / Wenchang Zhou / Jooyeon Lee / Ho Jeong Choi / Min Kim / José D Faraldo-Gómez / Richard K Hite /   Abstract: Structures of the human lysosomal K channel transmembrane protein 175 (TMEM175) in open and closed states revealed a novel architecture lacking the canonical K selectivity filter motif present in ...Structures of the human lysosomal K channel transmembrane protein 175 (TMEM175) in open and closed states revealed a novel architecture lacking the canonical K selectivity filter motif present in previously known K channel structures. A hydrophobic constriction composed of four isoleucine residues was resolved in the pore and proposed to serve as the gate in the closed state, and to confer ion selectivity in the open state. Here, we achieve higher-resolution structures of the open and closed states and employ molecular dynamics simulations to analyze the conducting properties of the putative open state, demonstrating that it is permeable to K and, to a lesser degree, also Na. Both cations must dehydrate significantly to penetrate the narrow hydrophobic constriction, but ion flow is assisted by a favorable electrostatic field generated by the protein that spans the length of the pore. The balance of these opposing energetic factors explains why permeation is feasible, and why TMEM175 is selective for K over Na, despite the absence of the canonical selectivity filter. Accordingly, mutagenesis experiments reveal an exquisite sensitivity of the channel to perturbations that mitigate the constriction. Together, these data reveal a novel mechanism for selective permeation of ions by TMEM175 that is unlike that of other K channels. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26627.map.gz emd_26627.map.gz | 107.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26627-v30.xml emd-26627-v30.xml emd-26627.xml emd-26627.xml | 16.9 KB 16.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_26627.png emd_26627.png | 98.5 KB | ||
| Filedesc metadata |  emd-26627.cif.gz emd-26627.cif.gz | 6 KB | ||
| Others |  emd_26627_half_map_1.map.gz emd_26627_half_map_1.map.gz emd_26627_half_map_2.map.gz emd_26627_half_map_2.map.gz | 200.3 MB 200.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26627 http://ftp.pdbj.org/pub/emdb/structures/EMD-26627 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26627 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26627 | HTTPS FTP |
-Validation report
| Summary document |  emd_26627_validation.pdf.gz emd_26627_validation.pdf.gz | 721.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_26627_full_validation.pdf.gz emd_26627_full_validation.pdf.gz | 721.2 KB | Display | |
| Data in XML |  emd_26627_validation.xml.gz emd_26627_validation.xml.gz | 15.7 KB | Display | |
| Data in CIF |  emd_26627_validation.cif.gz emd_26627_validation.cif.gz | 18.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26627 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26627 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26627 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26627 | HTTPS FTP |
-Related structure data
| Related structure data |  7unmMC  7unlC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_26627.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26627.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Closed TMEM175 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.85 Å | ||||||||||||||||||||||||||||||||||||
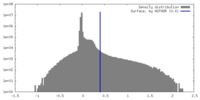
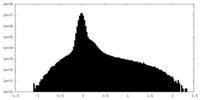
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half map A
| File | emd_26627_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
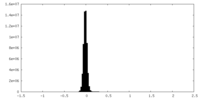
| Density Histograms |
-Half map: Half map B
| File | emd_26627_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
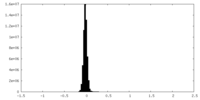
| Density Histograms |
- Sample components
Sample components
-Entire : TMEM175
| Entire | Name: TMEM175 |
|---|---|
| Components |
|
-Supramolecule #1: TMEM175
| Supramolecule | Name: TMEM175 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Endosomal/lysosomal potassium channel TMEM175
| Macromolecule | Name: Endosomal/lysosomal potassium channel TMEM175 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 55.667219 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MSQPRTPEQA LDTPGDCPPG RRDEDAGEGI QCSQRMLSFS DALLSIIATV MILPVTHTEI SPEQQFDRSV QRLLATRIAV YLMTFLIVT VAWAAHTRLF QVVGKTDDTL ALLNLACMMT ITFLPYTFSL MVTFPDVPLG IFLFCVCVIA IGVVQALIVG Y AFHFPHLL ...String: MSQPRTPEQA LDTPGDCPPG RRDEDAGEGI QCSQRMLSFS DALLSIIATV MILPVTHTEI SPEQQFDRSV QRLLATRIAV YLMTFLIVT VAWAAHTRLF QVVGKTDDTL ALLNLACMMT ITFLPYTFSL MVTFPDVPLG IFLFCVCVIA IGVVQALIVG Y AFHFPHLL SPQIQRSAHR ALYRRHVLGI VLQGPALCFA AAIFSLFFVP LSYLLMVTVI LLPYVSKVTG WCRDRLLGHR EP SAHPVEV FSFDLHEPLS KERVEAFSDG VYAIVATLLI LDICEDNVPD PKDVKERFSG SLVAALSATG PRFLAYFGSF ATV GLLWFA HHSLFLHVRK ATRAMGLLNT LSLAFVGGLP LAYQQTSAFA RQPRDELERV RVSCTIIFLA SIFQLAMWTT ALLH QAETL QPSVWFGGRE HVLMFAKLAL YPCASLLAFA STCLLSRFSV GIFHLMQIAV PCAFLLLRLL VGLALATLRV LRGLA RPEH PPPAPTGQDD PQSQLLPAPC UniProtKB: Endosomal/lysosomal proton channel TMEM175 |
-Macromolecule #2: POTASSIUM ION
| Macromolecule | Name: POTASSIUM ION / type: ligand / ID: 2 / Number of copies: 5 / Formula: K |
|---|---|
| Molecular weight | Theoretical: 39.098 Da |
-Macromolecule #3: water
| Macromolecule | Name: water / type: ligand / ID: 3 / Number of copies: 55 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| |||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 400 | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 61.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)