+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Complex of UBE2O with NAP1L1 | ||||||||||||||||||
 Map data Map data | Sharpened map | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | Ubiquitylation / CYTOSOLIC PROTEIN | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationhistone chaperone activity / (E3-independent) E2 ubiquitin-conjugating enzyme / positive regulation of BMP signaling pathway / positive regulation of neural precursor cell proliferation / retrograde transport, endosome to Golgi / positive regulation of neurogenesis / ubiquitin conjugating enzyme activity / protein K63-linked ubiquitination / protein monoubiquitination / ubiquitin-protein transferase activity ...histone chaperone activity / (E3-independent) E2 ubiquitin-conjugating enzyme / positive regulation of BMP signaling pathway / positive regulation of neural precursor cell proliferation / retrograde transport, endosome to Golgi / positive regulation of neurogenesis / ubiquitin conjugating enzyme activity / protein K63-linked ubiquitination / protein monoubiquitination / ubiquitin-protein transferase activity / ubiquitin protein ligase activity / melanosome / nervous system development / Antigen processing: Ubiquitination & Proteasome degradation / nucleosome assembly / histone binding / DNA replication / nuclear body / positive regulation of cell population proliferation / chromatin binding / chromatin / RNA binding / nucleoplasm / ATP binding / membrane / nucleus / cytoplasm / cytosol Similarity search - Function | ||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||||||||
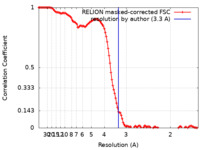
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | ||||||||||||||||||
 Authors Authors | Yip MCJ / Sedor SF / Shao S | ||||||||||||||||||
| Funding support |  United States, 5 items United States, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2022 Journal: Nat Struct Mol Biol / Year: 2022Title: Mechanism of client selection by the protein quality-control factor UBE2O. Authors: Matthew C J Yip / Samantha F Sedor / Sichen Shao /  Abstract: The E2/E3 enzyme UBE2O ubiquitylates diverse clients to mediate important processes, including targeting unassembled 'orphan' proteins for quality control and clearing ribosomes during erythropoiesis. ...The E2/E3 enzyme UBE2O ubiquitylates diverse clients to mediate important processes, including targeting unassembled 'orphan' proteins for quality control and clearing ribosomes during erythropoiesis. How quality-control factors, such as UBE2O, select clients on the basis of heterogeneous features is largely unknown. Here, we show that UBE2O client selection is regulated by ubiquitin binding and a cofactor, NAP1L1. Attaching a single ubiquitin onto a client enhances UBE2O binding and multi-mono-ubiquitylation. UBE2O also repurposes the histone chaperone NAP1L1 as an adapter to recruit a subset of clients. Cryo-EM structures of human UBE2O in complex with NAP1L1 reveal a malleable client recruitment interface that is autoinhibited by the intrinsically reactive UBC domain. Adding a ubiquitylated client identifies a distinct ubiquitin-binding SH3-like domain required for client selection. Our findings reveal how multivalency and a feed-forward mechanism drive the selection of protein quality-control clients. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26614.map.gz emd_26614.map.gz | 115.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26614-v30.xml emd-26614-v30.xml emd-26614.xml emd-26614.xml | 20.9 KB 20.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_26614_fsc.xml emd_26614_fsc.xml | 12.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_26614.png emd_26614.png | 100.8 KB | ||
| Filedesc metadata |  emd-26614.cif.gz emd-26614.cif.gz | 6.4 KB | ||
| Others |  emd_26614_additional_1.map.gz emd_26614_additional_1.map.gz emd_26614_additional_2.map.gz emd_26614_additional_2.map.gz emd_26614_half_map_1.map.gz emd_26614_half_map_1.map.gz emd_26614_half_map_2.map.gz emd_26614_half_map_2.map.gz | 71.9 MB 108.5 MB 114.1 MB 114.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26614 http://ftp.pdbj.org/pub/emdb/structures/EMD-26614 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26614 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26614 | HTTPS FTP |
-Related structure data
| Related structure data |  7un6MC  7un3C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_26614.map.gz / Format: CCP4 / Size: 149.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26614.map.gz / Format: CCP4 / Size: 149.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.825 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Unsharpened map
| File | emd_26614_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: deepEMhancer sharpened map
| File | emd_26614_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | deepEMhancer sharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 1
| File | emd_26614_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 2
| File | emd_26614_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Complex of UBE2O with NAP1L1
| Entire | Name: Complex of UBE2O with NAP1L1 |
|---|---|
| Components |
|
-Supramolecule #1: Complex of UBE2O with NAP1L1
| Supramolecule | Name: Complex of UBE2O with NAP1L1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: (E3-independent) E2 ubiquitin-conjugating enzyme
| Macromolecule | Name: (E3-independent) E2 ubiquitin-conjugating enzyme / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: (E3-independent) E2 ubiquitin-conjugating enzyme |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 147.187188 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MASWSHPQFE KGAWSHPQFE KGSWSHPQFE KGPAGSENLY FQGSGIRDRT MADPAAPTPA APAPAQAPAP APEAVPAPAA APVPAPAPA SDSASGPSSD SGPEAGSQRL LFSHDLVSGR YRGSVHFGLV RLIHGEDSDS EGEEEGRGSS GCSEAGGAGH E EGRASPLR ...String: MASWSHPQFE KGAWSHPQFE KGSWSHPQFE KGPAGSENLY FQGSGIRDRT MADPAAPTPA APAPAQAPAP APEAVPAPAA APVPAPAPA SDSASGPSSD SGPEAGSQRL LFSHDLVSGR YRGSVHFGLV RLIHGEDSDS EGEEEGRGSS GCSEAGGAGH E EGRASPLR RGYVRVQWYP EGVKQHVKET KLKLEDRSVV PRDVVRHMRS TDSQCGTVID VNIDCAVKLI GTNCIIYPVN SK DLQHIWP FMYGDYIAYD CWLGKVYDLK NQIILKLSNG ARCSMNTEDG AKLYDVCPHV SDSGLFFDDS YGFYPGQVLI GPA KIFSSV QWLSGVKPVL STKSKFRVVV EEVQVVELKV TWITKSFCPG GTDSVSPPPS VITQENLGRV KRLGCFDHAQ RQLG ERCLY VFPAKVEPAK IAWECPEKNC AQGEGSMAKK VKRLLKKQVV RIMSCSPDTQ CSRDHSMEDP DKKGESKTKS EAESA SPEE TPDGSASPVE MQDEGAEEPH EAGEQLPPFL LKEGRDDRLH SAEQDADDEA ADDTDDTSSV TSSASSTTSS QSGSGT SRK KSIPLSIKNL KRKHKRKKNK ITRDFKPGDR VAVEVVTTMT SADVMWQDGS VECNIRSNDL FPVHHLDNNE FCPGDFV VD KRVQSCPDPA VYGVVQSGDH IGRTCMVKWF KLRPSGDDVE LIGEEEDVSV YDIADHPDFR FRTTDIVIRI GNTEDGAP H KEDEPSVGQV ARVDVSSKVE VVWADNSKTI ILPQHLYNIE SEIEESDYDS VEGSTSGASS DEWEDDSDSW ETDNGLVED EHPKIEEPPI PPLEQPVAPE DKGVVISEEA ATAAVQGAVA MAAPMAGLME KAGKDGPPKS FRELKEAIKI LESLKNMTVE QLLTGSPTS PTVEPEKPTR EKKFLDDIKK LQENLKKTLD NVAIVEEEKM EAVPDVERKE DKPEGQSPVK AEWPSETPVL C QQCGGKPG VTFTSAKGEV FSVLEFAPSN HSFKKIEFQP PEAKKFFSTV RKEMALLATS LPEGIMVKTF EDRMDLFSAL IK GPTRTPY EDGLYLFDIQ LPNIYPAVPP HFCYLSQCSG RLNPNLYDNG KVKVSLLGTW IGKGTERWTS KSSLLQVLIS IQG LILVNE PYYNEAGFDS DRGLQEGYEN SRCYNEMALI RVVQSMTQLV RRPPEVFEQE IRQHFSTGGW RLVNRIESWL ETHA LLEKA QALPNGVPKA SSSPEPPAVA ELSDSGQQEP EDGGPAPGEA SQGSDSEGGA QGLASASRDH TDQTSETAPD ASVPP SVKP KKRRKSYRSF LPEKSGYPDI GFPLFPLSKG FIKSIRGVLT QFRAALLEAG MPECTEDK UniProtKB: (E3-independent) E2 ubiquitin-conjugating enzyme |
-Macromolecule #2: Nucleosome assembly protein 1-like 1
| Macromolecule | Name: Nucleosome assembly protein 1-like 1 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 48.363898 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MDYKDHDGDY KDHDIDYKDD DDKAGSADID NKEQSELDQD LDDVEEVEEE ETGEETKLKA RQLTVQMMQN PQILAALQER LDGLVETPT GYIESLPRVV KRRVNALKNL QVKCAQIEAK FYEEVHDLER KYAVLYQPLF DKRFEIINAI YEPTEEECEW K PDEEDEIS ...String: MDYKDHDGDY KDHDIDYKDD DDKAGSADID NKEQSELDQD LDDVEEVEEE ETGEETKLKA RQLTVQMMQN PQILAALQER LDGLVETPT GYIESLPRVV KRRVNALKNL QVKCAQIEAK FYEEVHDLER KYAVLYQPLF DKRFEIINAI YEPTEEECEW K PDEEDEIS EELKEKAKIE DEKKDEEKED PKGIPEFWLT VFKNVDLLSD MVQEHDEPIL KHLKDIKVKF SDAGQPMSFV LE FHFEPNE YFTNEVLTKT YRMRSEPDDS DPFSFDGPEI MGCTGCQIDW KKGKNVTLKT IKKKQKHKGR GTVRTVTKTV SND SFFNFF APPEVPESGD LDDDAEAILA ADFEIGHFLR ERIIPRSVLY FTGEAIEDDD DDYDEEGEEA DEEGEEEGDE ENDP DYDPK KDQNPAECKQ Q UniProtKB: Nucleosome assembly protein 1-like 1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 57.4 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.4000000000000001 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)