[English] 日本語
 Yorodumi
Yorodumi- EMDB-26600: SARS-CoV-2 Omicron-BA.1 2-RBD up Spike Protein Trimer without the... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | SARS-CoV-2 Omicron-BA.1 2-RBD up Spike Protein Trimer without the P986-P987 stabilizing mutations (S-GSAS-Omicron-BA.1) | |||||||||
 Map data Map data | Sharp map from homogeneous C1 refinement in cryosparc | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Omicron Spike protein / SARS-CoV-2 / variant of concern / 1-down / VIRAL PROTEIN / BA.1 | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.96 Å | |||||||||
 Authors Authors | Stalls V / Acharya P | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: bioRxiv / Year: 2022 Journal: bioRxiv / Year: 2022Title: Structural diversity of the SARS-CoV-2 Omicron spike. Authors: Sophie M-C Gobeil / Rory Henderson / Victoria Stalls / Katarzyna Janowska / Xiao Huang / Aaron May / Micah Speakman / Esther Beaudoin / Kartik Manne / Dapeng Li / Rob Parks / Maggie Barr / ...Authors: Sophie M-C Gobeil / Rory Henderson / Victoria Stalls / Katarzyna Janowska / Xiao Huang / Aaron May / Micah Speakman / Esther Beaudoin / Kartik Manne / Dapeng Li / Rob Parks / Maggie Barr / Margaret Deyton / Mitchell Martin / Katayoun Mansouri / Robert J Edwards / Gregory D Sempowski / Kevin O Saunders / Kevin Wiehe / Wilton Williams / Bette Korber / Barton F Haynes / Priyamvada Acharya Abstract: Aided by extensive spike protein mutation, the SARS-CoV-2 Omicron variant overtook the previously dominant Delta variant. Spike conformation plays an essential role in SARS-CoV-2 evolution via ...Aided by extensive spike protein mutation, the SARS-CoV-2 Omicron variant overtook the previously dominant Delta variant. Spike conformation plays an essential role in SARS-CoV-2 evolution via changes in receptor binding domain (RBD) and neutralizing antibody epitope presentation affecting virus transmissibility and immune evasion. Here, we determine cryo-EM structures of the Omicron and Delta spikes to understand the conformational impacts of mutations in each. The Omicron spike structure revealed an unusually tightly packed RBD organization with long range impacts that were not observed in the Delta spike. Binding and crystallography revealed increased flexibility at the functionally critical fusion peptide site in the Omicron spike. These results reveal a highly evolved Omicron spike architecture with possible impacts on its high levels of immune evasion and transmissibility. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26600.map.gz emd_26600.map.gz | 96.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26600-v30.xml emd-26600-v30.xml emd-26600.xml emd-26600.xml | 15.3 KB 15.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_26600.png emd_26600.png | 79.9 KB | ||
| Filedesc metadata |  emd-26600.cif.gz emd-26600.cif.gz | 5.6 KB | ||
| Others |  emd_26600_half_map_1.map.gz emd_26600_half_map_1.map.gz emd_26600_half_map_2.map.gz emd_26600_half_map_2.map.gz | 95.4 MB 95.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26600 http://ftp.pdbj.org/pub/emdb/structures/EMD-26600 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26600 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26600 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_26600.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26600.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharp map from homogeneous C1 refinement in cryosparc | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half A map from homogeneous C1 refinement in cryosparc
| File | emd_26600_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half A map from homogeneous C1 refinement in cryosparc | ||||||||||||
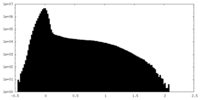
| Projections & Slices |
| ||||||||||||
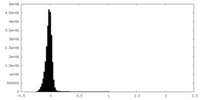
| Density Histograms |
-Half map: Half B map from homogeneous C1 refinement in cryosparc
| File | emd_26600_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half B map from homogeneous C1 refinement in cryosparc | ||||||||||||
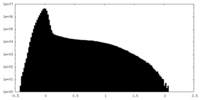
| Projections & Slices |
| ||||||||||||
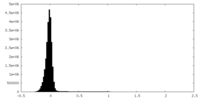
| Density Histograms |
- Sample components
Sample components
-Entire : SARS-CoV-2 S-GSAS-Omicron-BA.1 Spike Ectodomain
| Entire | Name: SARS-CoV-2 S-GSAS-Omicron-BA.1 Spike Ectodomain |
|---|---|
| Components |
|
-Supramolecule #1: SARS-CoV-2 S-GSAS-Omicron-BA.1 Spike Ectodomain
| Supramolecule | Name: SARS-CoV-2 S-GSAS-Omicron-BA.1 Spike Ectodomain / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 427 KDa |
-Macromolecule #1: Spike glycoprotein
| Macromolecule | Name: Spike glycoprotein / type: other / ID: 1 / Classification: other |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MFVFLVLLPL VSSQCVNLTT RTQLPPAYTN SFTRGVYYPD KVFRSSVLHS TQDLFLPFFS NVTWFHVISG TNGTKRFDNP VLPFNDGVYF ASIEKSNIIR GWIFGTTLDS KTQSLLIVNN ATNVVIKVCE FQFCNDPFLD HKNNKSWMES EFRVYSSANN CTFEYVSQPF ...String: MFVFLVLLPL VSSQCVNLTT RTQLPPAYTN SFTRGVYYPD KVFRSSVLHS TQDLFLPFFS NVTWFHVISG TNGTKRFDNP VLPFNDGVYF ASIEKSNIIR GWIFGTTLDS KTQSLLIVNN ATNVVIKVCE FQFCNDPFLD HKNNKSWMES EFRVYSSANN CTFEYVSQPF LMDLEGKQGN FKNLREFVFK NIDGYFKIYS KHTPIIVREP EDLPQGFSAL EPLVDLPIGI NITRFQTLLA LHRSYLTPGD SSSGWTAGAA AYYVGYLQPR TFLLKYNENG TITDAVDCAL DPLSETKCTL KSFTVEKGIY QTSNFRVQPT ESIVRFPNIT NLCPFDEVFN ATRFASVYAW NRKRISNCVA DYSVLYNLAP FFTFKCYGVS PTKLNDLCFT NVYADSFVIR GDEVRQIAPG QTGNIADYNY KLPDDFTGCV IAWNSNKLDS KVSGNYNYLY RLFRKSNLKP FERDISTEIY QAGNKPCNGV AGFNCYFPLR SYSFRPTYGV GHQPYRVVVL SFELLHAPAT VCGPKKSTNL VKNKCVNFNF NGLKGTGVLT ESNKKFLPFQ QFGRDIADTT DAVRDPQTLE ILDITPCSFG GVSVITPGTN TSNQVAVLYQ GVNCTEVPVA IHADQLTPTW RVYSTGSNVF QTRAGCLIGA EYVNNSYECD IPIGAGICAS YQTQTKSHGS ASSVASQSII AYTMSLGAEN SVAYSNNSIA IPTNFTISVT TEILPVSMTK TSVDCTMYIC GDSTECSNLL LQYGSFCTQL KRALTGIAVE QDKNTQEVFA QVKQIYKTPP IKYFGGFNFS QILPDPSKPS KRSFIEDLLF NKVTLADAGF IKQYGDCLGD IAARDLICAQ KFKGLTVLPP LLTDEMIAQY TSALLAGTIT SGWTFGAGAA LQIPFAMQMA YRFNGIGVTQ NVLYENQKLI ANQFNSAIGK IQDSLSSTAS ALGKLQDVVN HNAQALNTLV KQLSSKFGAI SSVLNDIFSR LDKVEAEVQI DRLITGRLQS LQTYVTQQLI RAAEIRASAN LAATKMSECV LGQSKRVDFC GKGYHLMSFP QSAPHGVVFL HVTYVPAQEK NFTTAPAICH DGKAHFPREG VFVSNGTHWF VTQRNFYEPQ IITTDNTFVS GNCDVVIGIV NNTVYDPLQP ELDS |
| Recombinant expression | Organism: Mammalia (mammals) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.5 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: NITROGEN |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.7000000000000001 µm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.96 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 101363 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)