[English] 日本語
 Yorodumi
Yorodumi- EMDB-26598: CryoEM structure of Go-coupled 5-HT5AR in complex with Lisuride -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of Go-coupled 5-HT5AR in complex with Lisuride | |||||||||
 Map data Map data | primary map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GPCR / Lisuride / active state / MEMBRANE PROTEIN / 5-HT5AR / HTR5A / Go | |||||||||
| Function / homology |  Function and homology information Function and homology informationGi/o-coupled serotonin receptor activity / Serotonin receptors / serotonin receptor activity / G protein-coupled serotonin receptor activity / neurotransmitter receptor activity / postsynaptic specialization membrane / G protein-coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger / adenylate cyclase-inhibiting serotonin receptor signaling pathway / hippocampus development / Olfactory Signaling Pathway ...Gi/o-coupled serotonin receptor activity / Serotonin receptors / serotonin receptor activity / G protein-coupled serotonin receptor activity / neurotransmitter receptor activity / postsynaptic specialization membrane / G protein-coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger / adenylate cyclase-inhibiting serotonin receptor signaling pathway / hippocampus development / Olfactory Signaling Pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / adenylate cyclase-activating G protein-coupled receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G-protein activation / Prostacyclin signalling through prostacyclin receptor / G beta:gamma signalling through CDC42 / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / photoreceptor disc membrane / Sensory perception of sweet, bitter, and umami (glutamate) taste / Glucagon-type ligand receptors / Adrenaline,noradrenaline inhibits insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (z) signalling events / cellular response to catecholamine stimulus / ADP signalling through P2Y purinoceptor 1 / ADORA2B mediated anti-inflammatory cytokines production / G beta:gamma signalling through PI3Kgamma / adenylate cyclase-activating dopamine receptor signaling pathway / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / GPER1 signaling / response to estradiol / Inactivation, recovery and regulation of the phototransduction cascade / cellular response to prostaglandin E stimulus / G-protein beta-subunit binding / heterotrimeric G-protein complex / G alpha (12/13) signalling events / sensory perception of taste / extracellular vesicle / signaling receptor complex adaptor activity / Thrombin signalling through proteinase activated receptors (PARs) / retina development in camera-type eye / GTPase binding / Ca2+ pathway / fibroblast proliferation / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / perikaryon / G alpha (i) signalling events / G alpha (s) signalling events / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (q) signalling events / chemical synaptic transmission / Ras protein signal transduction / Extra-nuclear estrogen signaling / cell population proliferation / G protein-coupled receptor signaling pathway / lysosomal membrane / GTPase activity / dendrite / synapse / protein-containing complex binding / signal transduction / extracellular exosome / membrane / plasma membrane / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
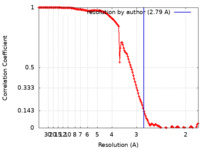
| Method | single particle reconstruction / cryo EM / Resolution: 2.79 Å | |||||||||
 Authors Authors | Zhang S / Fay JF / Roth BL | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2022 Journal: Nat Struct Mol Biol / Year: 2022Title: Inactive and active state structures template selective tools for the human 5-HT receptor. Authors: Shicheng Zhang / He Chen / Chengwei Zhang / Ying Yang / Petr Popov / Jing Liu / Brian E Krumm / Can Cao / Kuglae Kim / Yan Xiong / Vsevolod Katritch / Brian K Shoichet / Jian Jin / Jonathan ...Authors: Shicheng Zhang / He Chen / Chengwei Zhang / Ying Yang / Petr Popov / Jing Liu / Brian E Krumm / Can Cao / Kuglae Kim / Yan Xiong / Vsevolod Katritch / Brian K Shoichet / Jian Jin / Jonathan F Fay / Bryan L Roth /   Abstract: Serotonin receptors are important targets for established therapeutics and drug development as they are expressed throughout the human body and play key roles in cell signaling. There are 12 ...Serotonin receptors are important targets for established therapeutics and drug development as they are expressed throughout the human body and play key roles in cell signaling. There are 12 serotonergic G protein-coupled receptor members encoded in the human genome, of which the 5-hydroxytryptamine (5-HT) receptor (5-HTR) is the least understood and lacks selective tool compounds. Here, we report four high-resolution (2.73-2.80 Å) structures of human 5-HTRs, including an inactive state structure bound to an antagonist AS2674723 by crystallization and active state structures bound to a partial agonist lisuride and two full agonists, 5-carboxamidotryptamine (5-CT) and methylergometrine, by cryo-EM. Leveraging the new structures, we developed a highly selective and potent antagonist for 5-HTR. Collectively, these findings both enhance our understanding of this enigmatic receptor and provide a roadmap for structure-based drug discovery for 5-HTR. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26598.map.gz emd_26598.map.gz | 54.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26598-v30.xml emd-26598-v30.xml emd-26598.xml emd-26598.xml | 24.5 KB 24.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_26598_fsc.xml emd_26598_fsc.xml | 13.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_26598.png emd_26598.png | 67.2 KB | ||
| Masks |  emd_26598_msk_1.map emd_26598_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-26598.cif.gz emd-26598.cif.gz | 7.2 KB | ||
| Others |  emd_26598_half_map_1.map.gz emd_26598_half_map_1.map.gz emd_26598_half_map_2.map.gz emd_26598_half_map_2.map.gz | 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26598 http://ftp.pdbj.org/pub/emdb/structures/EMD-26598 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26598 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26598 | HTTPS FTP |
-Validation report
| Summary document |  emd_26598_validation.pdf.gz emd_26598_validation.pdf.gz | 814.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_26598_full_validation.pdf.gz emd_26598_full_validation.pdf.gz | 813.7 KB | Display | |
| Data in XML |  emd_26598_validation.xml.gz emd_26598_validation.xml.gz | 18.3 KB | Display | |
| Data in CIF |  emd_26598_validation.cif.gz emd_26598_validation.cif.gz | 24.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26598 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26598 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26598 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26598 | HTTPS FTP |
-Related structure data
| Related structure data |  7um6MC  7um4C  7um5C  7um7C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
| EM raw data |  EMPIAR-11036 (Title: CryoEM structure of Go-coupled 5-HT5AR in complex with Lisuride EMPIAR-11036 (Title: CryoEM structure of Go-coupled 5-HT5AR in complex with LisurideData size: 406.8 Data #1: Aligned multi-frame micrographs [micrographs - single frame]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_26598.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26598.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | primary map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.91 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_26598_msk_1.map emd_26598_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map A
| File | emd_26598_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map B
| File | emd_26598_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Go-coupled 5-HT5AR complex
| Entire | Name: Go-coupled 5-HT5AR complex |
|---|---|
| Components |
|
-Supramolecule #1: Go-coupled 5-HT5AR complex
| Supramolecule | Name: Go-coupled 5-HT5AR complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|
-Supramolecule #2: 5-hydroxytryptamine receptor 5A, miniGo protein, Guanine nucleoti...
| Supramolecule | Name: 5-hydroxytryptamine receptor 5A, miniGo protein, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #3: Single-chain variable fragment scFv16
| Supramolecule | Name: Single-chain variable fragment scFv16 / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #5 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: 5-hydroxytryptamine receptor 5A
| Macromolecule | Name: 5-hydroxytryptamine receptor 5A / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 36.884168 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SSPLLSVFGV LILTLLGFLV AATFAWNLLV LATILRVRTF HRVPHNLVAS MAVSDVLVAA LVMPLSLVHE LSGRRWQLGR RLCQLWIAC DVLCCTASIW NVTAIALDRY WSITRPMEYT LRTRKCVSNV MIALTWALSA VISLAPLLFG WGETYSEGSE E CQVSREPS ...String: SSPLLSVFGV LILTLLGFLV AATFAWNLLV LATILRVRTF HRVPHNLVAS MAVSDVLVAA LVMPLSLVHE LSGRRWQLGR RLCQLWIAC DVLCCTASIW NVTAIALDRY WSITRPMEYT LRTRKCVSNV MIALTWALSA VISLAPLLFG WGETYSEGSE E CQVSREPS YAVFSTVGAF YLPLCVVLFV YWKIYKAAKF RVGSRKTNSV SPISEAVEVK DSAKQPQMVF TVRHATVTFQ PE GDTWREQ KEQRAALMVG ILIGVFVLCW IPFFLTELIS PLCSCDIPAI WKSIFLWLGY SNSFFNPLIY TAFNKNYNSA FKN FFSRQH UniProtKB: 5-hydroxytryptamine receptor 5A |
-Macromolecule #2: miniGo protein
| Macromolecule | Name: miniGo protein / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 25.159777 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: TLSAEDKAAV ERSKMIEKNL KEDGISAAKD VKLLLLGADN SGKSTIVKQM KIIHGGSGGS GGTTGIVETH FTFKNLHFRL FDVGGQRSE RKKWIHCFED VTAIIFCVDL SDYNRMHESL MLFDSICNNK FFIDTSIILF LNKKDLFGEK IKKSPLTICF P EYTGPNTY ...String: TLSAEDKAAV ERSKMIEKNL KEDGISAAKD VKLLLLGADN SGKSTIVKQM KIIHGGSGGS GGTTGIVETH FTFKNLHFRL FDVGGQRSE RKKWIHCFED VTAIIFCVDL SDYNRMHESL MLFDSICNNK FFIDTSIILF LNKKDLFGEK IKKSPLTICF P EYTGPNTY EDAAAYIQAQ FESKNRSPNK EIYCHMTCAT DTNNAQVIFD AVTDIIIANN LRGCGLY |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 37.285734 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SELDQLRQEA EQLKNQIRDA RKACADATLS QITNNIDPVG RIQMRTRRTL RGHLAKIYAM HWGTDSRLLV SASQDGKLII WDSYTTNKV HAIPLRSSWV MTCAYAPSGN YVACGGLDNI CSIYNLKTRE GNVRVSRELA GHTGYLSCCR FLDDNQIVTS S GDTTCALW ...String: SELDQLRQEA EQLKNQIRDA RKACADATLS QITNNIDPVG RIQMRTRRTL RGHLAKIYAM HWGTDSRLLV SASQDGKLII WDSYTTNKV HAIPLRSSWV MTCAYAPSGN YVACGGLDNI CSIYNLKTRE GNVRVSRELA GHTGYLSCCR FLDDNQIVTS S GDTTCALW DIETGQQTTT FTGHTGDVMS LSLAPDTRLF VSGACDASAK LWDVREGMCR QTFTGHESDI NAICFFPNGN AF ATGSDDA TCRLFDLRAD QELMTYSHDN IICGITSVSF SKSGRLLLAG YDDFNCNVWD ALKADRAGVL AGHDNRVSCL GVT DDGMAV ATGSWDSFLK IWN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #4: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.861143 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFCAI L UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #5: Single-chain variable fragment scFv16
| Macromolecule | Name: Single-chain variable fragment scFv16 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 26.679721 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DVQLVESGGG LVQPGGSRKL SCSASGFAFS SFGMHWVRQA PEKGLEWVAY ISSGSGTIYY ADTVKGRFTI SRDDPKNTLF LQMTSLRSE DTAMYYCVRS IYYYGSSPFD FWGQGTTLTV SSGGGGSGGG GSGGGGSDIV MTQATSSVPV TPGESVSISC R SSKSLLHS ...String: DVQLVESGGG LVQPGGSRKL SCSASGFAFS SFGMHWVRQA PEKGLEWVAY ISSGSGTIYY ADTVKGRFTI SRDDPKNTLF LQMTSLRSE DTAMYYCVRS IYYYGSSPFD FWGQGTTLTV SSGGGGSGGG GSGGGGSDIV MTQATSSVPV TPGESVSISC R SSKSLLHS NGNTYLYWFL QRPGQSPQLL IYRMSNLASG VPDRFSGSGS GTAFTLTISR LEAEDVGVYY CMQHLEYPLT FG AGTKLEL KAAA |
-Macromolecule #6: N,N-diethyl-N'-[(8alpha)-6-methyl-9,10-didehydroergolin-8-yl]urea
| Macromolecule | Name: N,N-diethyl-N'-[(8alpha)-6-methyl-9,10-didehydroergolin-8-yl]urea type: ligand / ID: 6 / Number of copies: 1 / Formula: H8G |
|---|---|
| Molecular weight | Theoretical: 338.447 Da |
| Chemical component information | 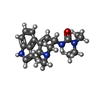 ChemComp-H8G: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI 10 |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 43.8 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.211 µm / Nominal defocus min: 0.14300000000000002 µm |
 Movie
Movie Controller
Controller


























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)