[English] 日本語
 Yorodumi
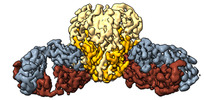
Yorodumi- EMDB-26594: Lineage I (Pinneo) Lassa virus glycoprotein bound to 18.5C-M30 Fab -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Lineage I (Pinneo) Lassa virus glycoprotein bound to 18.5C-M30 Fab | ||||||||||||
 Map data Map data | Sharpened map | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | glycoprotein / antibody / lassa virus / VIRAL PROTEIN / VIRAL PROTEIN-IMMUNE SYSTEM complex | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationhost cell Golgi membrane / receptor-mediated endocytosis of virus by host cell / host cell endoplasmic reticulum membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / host cell plasma membrane / virion membrane / metal ion binding / membrane Similarity search - Function | ||||||||||||
| Biological species |  Lassa mammarenavirus / Lassa mammarenavirus /  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
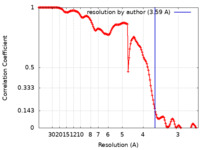
| Method | single particle reconstruction / cryo EM / Resolution: 3.59 Å | ||||||||||||
 Authors Authors | Buck TK / Enriquez AS / Hastie KM | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: mBio / Year: 2022 Journal: mBio / Year: 2022Title: Neutralizing Antibodies against Lassa Virus Lineage I. Authors: Tierra K Buck / Adrian S Enriquez / Sharon L Schendel / Michelle A Zandonatti / Stephanie S Harkins / Haoyang Li / Alex Moon-Walker / James E Robinson / Luis M Branco / Robert F Garry / ...Authors: Tierra K Buck / Adrian S Enriquez / Sharon L Schendel / Michelle A Zandonatti / Stephanie S Harkins / Haoyang Li / Alex Moon-Walker / James E Robinson / Luis M Branco / Robert F Garry / Erica Ollmann Saphire / Kathryn M Hastie /  Abstract: Lassa virus (LASV) is the causative agent of the deadly Lassa fever (LF). Seven distinct LASV lineages circulate through western Africa, among which lineage I (LI), the first to be identified, is ...Lassa virus (LASV) is the causative agent of the deadly Lassa fever (LF). Seven distinct LASV lineages circulate through western Africa, among which lineage I (LI), the first to be identified, is particularly resistant to antibody neutralization. Lineage I LASV evades neutralization by half of known antibodies in the GPC-A antibody competition group and all but one of the antibodies in the GPC-B competition group. Here, we solve two cryo-electron microscopy (cryo-EM) structures of LI GP in complex with a GPC-A and a GPC-B antibody. We used complementary structural and biochemical techniques to identify single-amino-acid substitutions in LI that are responsible for immune evasion by each antibody group. Further, we show that LI infection is more dependent on the endosomal receptor lysosome-associated membrane protein 1 (LAMP1) for viral entry relative to LIV. In the absence of LAMP1, LI requires a more acidic fusion pH to initiate membrane fusion with the host cell relative to LIV. No vaccine or therapeutics are approved to prevent LASV infection or treat LF. All vaccine platforms currently under development present only the LIV GP sequence. However, our data suggest that the high genetic diversity of LASV may be problematic for designing both a broadly reactive immunogen and therapeutic. Here, we examine antibodies that are highly potent against LIV yet are ineffective against LI. By pinpointing LI mutations responsible for this decrease in antibody efficacy, we suggest that future vaccine platforms may need to incorporate specific LI-like mutations in order to generate a broadly neutralizing antibody response against all LASV lineages. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26594.map.gz emd_26594.map.gz | 244.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26594-v30.xml emd-26594-v30.xml emd-26594.xml emd-26594.xml | 20.8 KB 20.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_26594_fsc.xml emd_26594_fsc.xml | 14.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_26594.png emd_26594.png | 100.2 KB | ||
| Filedesc metadata |  emd-26594.cif.gz emd-26594.cif.gz | 6.2 KB | ||
| Others |  emd_26594_additional_1.map.gz emd_26594_additional_1.map.gz emd_26594_half_map_1.map.gz emd_26594_half_map_1.map.gz emd_26594_half_map_2.map.gz emd_26594_half_map_2.map.gz | 138 MB 255.2 MB 255.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26594 http://ftp.pdbj.org/pub/emdb/structures/EMD-26594 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26594 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26594 | HTTPS FTP |
-Related structure data
| Related structure data |  7ul7MC  7udsC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_26594.map.gz / Format: CCP4 / Size: 274.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26594.map.gz / Format: CCP4 / Size: 274.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
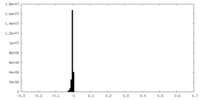
| Annotation | Sharpened map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.296 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Unsharpened map
| File | emd_26594_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
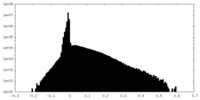
| Annotation | Unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map A
| File | emd_26594_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
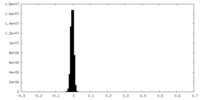
| Annotation | Half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_26594_half_map_2.map | ||||||||||||
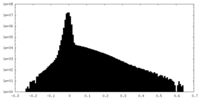
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Lineage I (Pinneo) Lassa virus glycoprotein bound to 18.5C-M30 Fab
| Entire | Name: Lineage I (Pinneo) Lassa virus glycoprotein bound to 18.5C-M30 Fab |
|---|---|
| Components |
|
-Supramolecule #1: Lineage I (Pinneo) Lassa virus glycoprotein bound to 18.5C-M30 Fab
| Supramolecule | Name: Lineage I (Pinneo) Lassa virus glycoprotein bound to 18.5C-M30 Fab type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Molecular weight | Theoretical: 350 KDa |
-Macromolecule #1: Glycoprotein G1
| Macromolecule | Name: Glycoprotein G1 / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Lassa mammarenavirus Lassa mammarenavirus |
| Molecular weight | Theoretical: 28.987398 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGQIITFFQE VPHVIEEVMN IVLIALSLLA ILKGLYNIAT CGIIGLVAFL FLCGKSCSLT LKGGYELQTL ELNMETLNMT MPLSCTKNS SHHYIRVGNE TGLELTLTNT SIINHKFCNL SDAHKKNLYD HALMSIISTF HLSIPNFNQY EAMSCDFNGG K ISVQYNLS ...String: MGQIITFFQE VPHVIEEVMN IVLIALSLLA ILKGLYNIAT CGIIGLVAFL FLCGKSCSLT LKGGYELQTL ELNMETLNMT MPLSCTKNS SHHYIRVGNE TGLELTLTNT SIINHKFCNL SDAHKKNLYD HALMSIISTF HLSIPNFNQY EAMSCDFNGG K ISVQYNLS HSYAGDAAEH CGTVANGVLQ TFMRMAWGGR YIALDSGCGN WDCIMTSYQY LIIQNTTWED HCQFSRPSPI GY LGLLSQR TRDIYISRRR R UniProtKB: Pre-glycoprotein polyprotein GP complex |
-Macromolecule #2: 18.5C-M30 Fab Light Chain
| Macromolecule | Name: 18.5C-M30 Fab Light Chain / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 25.880822 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: METDTLLLWV LLLWVPGSTG DDIVLTQSPG TLSLSPGERA TLSCRASQSV ISYYVAWYQH KGGQAPRLLI YGASSRATGV PDRFSGSGS GTDFTLTISS LEPEDFALYY CQYYGSSPLW AFGQGTKVEI KRTVAAPSVF IFPPSDEQLK SGTASVVCLL N NFYPREAK ...String: METDTLLLWV LLLWVPGSTG DDIVLTQSPG TLSLSPGERA TLSCRASQSV ISYYVAWYQH KGGQAPRLLI YGASSRATGV PDRFSGSGS GTDFTLTISS LEPEDFALYY CQYYGSSPLW AFGQGTKVEI KRTVAAPSVF IFPPSDEQLK SGTASVVCLL N NFYPREAK VQWKVDNALQ SGNSQESVTE QDSKDSTYSL SSTLTLSKAD YEKHKVYACE VTHQGLSSPV TKSFNRGEC |
-Macromolecule #3: 18.5C-M30 Fab Heavy Chain
| Macromolecule | Name: 18.5C-M30 Fab Heavy Chain / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 26.737285 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: METDTLLLWV LLLWVPGSTG DEVQLVQSGG GLVRPGGSLR LSCAAAGFTF KSYSMNWVRQ APGRGLEWVS SITSRGGSKT YYADVVKGR FTVSRDNAKQ SLYLQMNSLR AEDTAIYFCA RSRHSTSQPS YMDVWGRKIT VIVSSASTKG PSVFPLAPSS K STSGGTAA ...String: METDTLLLWV LLLWVPGSTG DEVQLVQSGG GLVRPGGSLR LSCAAAGFTF KSYSMNWVRQ APGRGLEWVS SITSRGGSKT YYADVVKGR FTVSRDNAKQ SLYLQMNSLR AEDTAIYFCA RSRHSTSQPS YMDVWGRKIT VIVSSASTKG PSVFPLAPSS K STSGGTAA LGCLVKDYFP EPVTVSWNSG ALTSGVHTFP AVLQSSGLYS LSSVVTVPSS SLGTQTYICN VNHKPSNTKV DK RVEPKSC DK |
-Macromolecule #4: Glycoprotein G2
| Macromolecule | Name: Glycoprotein G2 / type: protein_or_peptide / ID: 4 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Lassa mammarenavirus Lassa mammarenavirus |
| Molecular weight | Theoretical: 23.348145 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GTFTWTLSDS EGNETPGGYC LTRWMLIEAE LKCFGNTAVA KCNEKHDEEF CDMLRLFDFN KQAIRRLKAP AQMSIQLINK AVNALINDQ LIMKNHLRDI MCIPYCNYSK YWYLNHTSSG RTSLPKCWLI SNGSYLNETQ FSDDIEQQAD NMITEMLQKE Y LPETGLVD ...String: GTFTWTLSDS EGNETPGGYC LTRWMLIEAE LKCFGNTAVA KCNEKHDEEF CDMLRLFDFN KQAIRRLKAP AQMSIQLINK AVNALINDQ LIMKNHLRDI MCIPYCNYSK YWYLNHTSSG RTSLPKCWLI SNGSYLNETQ FSDDIEQQAD NMITEMLQKE Y LPETGLVD LEVDDDDKAG WSHPQFEKGG GSGGGSGGGS WSHPQFEK UniProtKB: Pre-glycoprotein polyprotein GP complex |
-Macromolecule #6: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 6 / Number of copies: 22 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 Component:
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)