+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of full-length dimeric ClbP | |||||||||
 Map data Map data | Final map for ClbP | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | colibactin peptidase / S12 peptidase / inner-membrane hydrolase / HYDROLASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationantibiotic catabolic process / beta-lactamase activity / beta-lactamase / outer membrane-bounded periplasmic space / response to antibiotic / membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
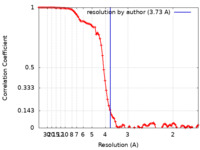
| Method | single particle reconstruction / cryo EM / Resolution: 3.73 Å | |||||||||
 Authors Authors | Velilla JA / Walsh Jr RM / Gaudet R | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Chem Biol / Year: 2023 Journal: Nat Chem Biol / Year: 2023Title: Structural basis of colibactin activation by the ClbP peptidase. Authors: José A Velilla / Matthew R Volpe / Grace E Kenney / Richard M Walsh / Emily P Balskus / Rachelle Gaudet /  Abstract: Colibactin, a DNA cross-linking agent produced by gut bacteria, is implicated in colorectal cancer. Its biosynthesis uses a prodrug resistance mechanism: a non-toxic precursor assembled in the ...Colibactin, a DNA cross-linking agent produced by gut bacteria, is implicated in colorectal cancer. Its biosynthesis uses a prodrug resistance mechanism: a non-toxic precursor assembled in the cytoplasm is activated after export to the periplasm. This activation is mediated by ClbP, an inner-membrane peptidase with an N-terminal periplasmic catalytic domain and a C-terminal three-helix transmembrane domain. Although the transmembrane domain is required for colibactin activation, its role in catalysis is unclear. Our structure of full-length ClbP bound to a product analog reveals an interdomain interface important for substrate binding and enzyme stability and interactions that explain the selectivity of ClbP for the N-acyl-D-asparagine prodrug motif. Based on structural and biochemical evidence, we propose that ClbP dimerizes to form an extended substrate-binding site that can accommodate a pseudodimeric precolibactin with its two terminal prodrug motifs in the two ClbP active sites, thus enabling the coordinated activation of both electrophilic warheads. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26593.map.gz emd_26593.map.gz | 59.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26593-v30.xml emd-26593-v30.xml emd-26593.xml emd-26593.xml | 18.6 KB 18.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_26593_fsc.xml emd_26593_fsc.xml | 8.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_26593.png emd_26593.png | 190 KB | ||
| Filedesc metadata |  emd-26593.cif.gz emd-26593.cif.gz | 6.6 KB | ||
| Others |  emd_26593_half_map_1.map.gz emd_26593_half_map_1.map.gz emd_26593_half_map_2.map.gz emd_26593_half_map_2.map.gz | 59.3 MB 59.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26593 http://ftp.pdbj.org/pub/emdb/structures/EMD-26593 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26593 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26593 | HTTPS FTP |
-Related structure data
| Related structure data |  7ul6MC  7mdeC  7mdfC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_26593.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26593.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Final map for ClbP | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.825 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half map A for ClbP
| File | emd_26593_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A for ClbP | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map B for ClbP
| File | emd_26593_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B for ClbP | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : ClbP
| Entire | Name: ClbP |
|---|---|
| Components |
|
-Supramolecule #1: ClbP
| Supramolecule | Name: ClbP / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all / Details: Full-length ClbP |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 110 KDa |
-Macromolecule #1: Beta-lactamase
| Macromolecule | Name: Beta-lactamase / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: beta-lactamase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 53.523309 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: QEHEPIGAQD ERLSTLIHQR MQEAKVPALS VSVTIKGVRQ RFVYGVADVA SQKANTLDTV YELGSMSKAF TGLVVQILIQ EGRLRQGDD IITYLPEMRL NYQGKPASLT VADFLYHTSG LPFSTLARLE NPMPGSAVAQ QLRNENLLFA PGAKFSYASA N YDVLGAVI ...String: QEHEPIGAQD ERLSTLIHQR MQEAKVPALS VSVTIKGVRQ RFVYGVADVA SQKANTLDTV YELGSMSKAF TGLVVQILIQ EGRLRQGDD IITYLPEMRL NYQGKPASLT VADFLYHTSG LPFSTLARLE NPMPGSAVAQ QLRNENLLFA PGAKFSYASA N YDVLGAVI ENVTGKTFTE VIAERLTQPL GMSATVAVKG DEIIVNKASG YKLGFGKPVL FHAPLARNHV PAAYIHSTLP DM EIWIDAW LHRKALPATL REAMSNSWRG NSDVPLAADN RILYASGWFI DQNQGPYISH GGQNPNFSSC IALRPDQQIG IVA LANMNS NLILQLCADI DNYLRIGKYA DGAGDAITAT DTLFVYLTLL LCFWGAVVVV RGAFRVYRAT AHGPGKQQRL RLRV RDYII ALAVPGLVAA MLYVAPGILS PGLDWRFILV WGPSSVLAIP FGIILLAFVL TLNHQIKRIL LHNKEWDDEH HHHHH HHHH UniProtKB: Beta-lactamase |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3.5 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.3 Component:
| ||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 400 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.039 kPa / Details: 30 s glow discharge at 15mA | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 295 K / Instrument: FEI VITROBOT MARK IV Details: Three uL of sample were deposited onto 400 mesh Quantifoil Cu 1.2/1.3 grids that had been glow discharged in a PELCO easiGLOW (Ted Pella) at 0.39 mBar, 15 mA for 30 s. Samples were vitrified ...Details: Three uL of sample were deposited onto 400 mesh Quantifoil Cu 1.2/1.3 grids that had been glow discharged in a PELCO easiGLOW (Ted Pella) at 0.39 mBar, 15 mA for 30 s. Samples were vitrified in 100% liquid ethane using a Vitrobot Mark IV (Thermo Fisher Scientific), with a wait time of 30 s, blot time of 5 s and a blot force of 16 at 100% humidity.. | ||||||||||||
| Details | Protein was purified by Ni affinity chromatography followed by SEC on an S200 10/300 column equilibrated with 10 mM HEPES pH 7.3, 200 mM NaCl, 0.06% GDN. Sample used for preparing grids came from the peak fraction and was not concentrated. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Number grids imaged: 1 / Number real images: 3888 / Average exposure time: 2.5 sec. / Average electron dose: 76.191 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated magnification: 60606 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)