+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of Human SAMHD1 with Non-Hydrolysable dGTP Analog | |||||||||
 Map data Map data | Human SAMHD1 with dGTP(alpha)S Nucleotide Final CryoEM Map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | dNTP Triphosphatase / HYDROLASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationNucleotide catabolism / Hydrolases; Acting on ester bonds; Triphosphoric-monoester hydrolases / deoxynucleoside triphosphate hydrolase activity / dGTP binding / dATP catabolic process / deoxyribonucleotide catabolic process / tetraspanin-enriched microdomain / dGTPase activity / dGTP catabolic process / DNA strand resection involved in replication fork processing ...Nucleotide catabolism / Hydrolases; Acting on ester bonds; Triphosphoric-monoester hydrolases / deoxynucleoside triphosphate hydrolase activity / dGTP binding / dATP catabolic process / deoxyribonucleotide catabolic process / tetraspanin-enriched microdomain / dGTPase activity / dGTP catabolic process / DNA strand resection involved in replication fork processing / negative regulation of type I interferon-mediated signaling pathway / regulation of innate immune response / RNA nuclease activity / somatic hypermutation of immunoglobulin genes / double-strand break repair via homologous recombination / Interferon alpha/beta signaling / single-stranded DNA binding / site of double-strand break / protein homotetramerization / defense response to virus / nucleic acid binding / immune response / innate immune response / DNA damage response / GTP binding / RNA binding / zinc ion binding / nucleoplasm / identical protein binding / nucleus / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.89 Å | |||||||||
 Authors Authors | Huynh KW / Ammirati M / Han S | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nucleic Acids Res / Year: 2022 Journal: Nucleic Acids Res / Year: 2022Title: Phosphorylation of SAMHD1 Thr592 increases C-terminal domain dynamics, tetramer dissociation and ssDNA binding kinetics. Authors: Benjamin Orris / Kevin W Huynh / Mark Ammirati / Seungil Han / Ben Bolaños / Jason Carmody / Matthew D Petroski / Benedikt Bosbach / David J Shields / James T Stivers /  Abstract: SAM and HD domain containing deoxynucleoside triphosphate triphosphohydrolase 1 (SAMHD1) is driven into its activated tetramer form by binding of GTP activator and dNTP activators/substrates. In ...SAM and HD domain containing deoxynucleoside triphosphate triphosphohydrolase 1 (SAMHD1) is driven into its activated tetramer form by binding of GTP activator and dNTP activators/substrates. In addition, the inactive monomeric and dimeric forms of the enzyme bind to single-stranded (ss) nucleic acids. During DNA replication SAMHD1 can be phosphorylated by CDK1 and CDK2 at its C-terminal threonine 592 (pSAMHD1), localizing the enzyme to stalled replication forks (RFs) to promote their restart. Although phosphorylation has only a small effect on the dNTPase activity and ssDNA binding affinity of SAMHD1, perturbation of the native T592 by phosphorylation decreased the thermal stability of tetrameric SAMHD1 and accelerated tetramer dissociation in the absence and presence of ssDNA (∼15-fold). In addition, we found that ssDNA binds competitively with GTP to the A1 site. A full-length SAMHD1 cryo-EM structure revealed substantial dynamics in the C-terminal domain (which contains T592), which could be modulated by phosphorylation. We propose that T592 phosphorylation increases tetramer dynamics and allows invasion of ssDNA into the A1 site and the previously characterized DNA binding surface at the dimer-dimer interface. These features are consistent with rapid and regiospecific inactivation of pSAMHD1 dNTPase at RFs or other sites of free ssDNA in cells. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26567.map.gz emd_26567.map.gz | 96.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26567-v30.xml emd-26567-v30.xml emd-26567.xml emd-26567.xml | 20.5 KB 20.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_26567_fsc.xml emd_26567_fsc.xml | 10.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_26567.png emd_26567.png | 79.9 KB | ||
| Filedesc metadata |  emd-26567.cif.gz emd-26567.cif.gz | 6.8 KB | ||
| Others |  emd_26567_half_map_1.map.gz emd_26567_half_map_1.map.gz emd_26567_half_map_2.map.gz emd_26567_half_map_2.map.gz | 79.4 MB 79.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26567 http://ftp.pdbj.org/pub/emdb/structures/EMD-26567 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26567 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26567 | HTTPS FTP |
-Validation report
| Summary document |  emd_26567_validation.pdf.gz emd_26567_validation.pdf.gz | 989.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_26567_full_validation.pdf.gz emd_26567_full_validation.pdf.gz | 988.8 KB | Display | |
| Data in XML |  emd_26567_validation.xml.gz emd_26567_validation.xml.gz | 17.5 KB | Display | |
| Data in CIF |  emd_26567_validation.cif.gz emd_26567_validation.cif.gz | 23 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26567 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26567 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26567 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26567 | HTTPS FTP |
-Related structure data
| Related structure data |  7ujnMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_26567.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26567.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Human SAMHD1 with dGTP(alpha)S Nucleotide Final CryoEM Map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.84 Å | ||||||||||||||||||||||||||||||||||||
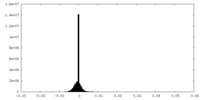
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Human SAMHD1 with dGTP(alpha)S Nucleotide Half Map 2
| File | emd_26567_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Human SAMHD1 with dGTP(alpha)S Nucleotide Half Map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
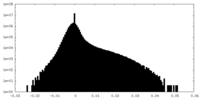
| Density Histograms |
-Half map: Human SAMHD1 with dGTP(alpha)S Nucleotide Half Map 1
| File | emd_26567_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Human SAMHD1 with dGTP(alpha)S Nucleotide Half Map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
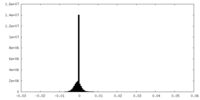
| Density Histograms |
- Sample components
Sample components
-Entire : Human SAMHD1 with Non-Hydrolysable dGTP Analog
| Entire | Name: Human SAMHD1 with Non-Hydrolysable dGTP Analog |
|---|---|
| Components |
|
-Supramolecule #1: Human SAMHD1 with Non-Hydrolysable dGTP Analog
| Supramolecule | Name: Human SAMHD1 with Non-Hydrolysable dGTP Analog / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 288 KDa |
-Macromolecule #1: Deoxynucleoside triphosphate triphosphohydrolase SAMHD1
| Macromolecule | Name: Deoxynucleoside triphosphate triphosphohydrolase SAMHD1 type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO EC number: Hydrolases; Acting on ester bonds; Triphosphoric-monoester hydrolases |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 72.305414 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MQRADSEQPS KRPRCDDSPR TPSNTPSAEA DWSPGLELHP DYKTWGPEQV CSFLRRGGFE EPVLLKNIRE NEITGALLPC LDESRFENL GVSSLGERKK LLSYIQRLVQ IHVDTMKVIN DPIHGHIELH PLLVRIIDTP QFQRLRYIKQ LGGGYYVFPG A SHNRFEHS ...String: MQRADSEQPS KRPRCDDSPR TPSNTPSAEA DWSPGLELHP DYKTWGPEQV CSFLRRGGFE EPVLLKNIRE NEITGALLPC LDESRFENL GVSSLGERKK LLSYIQRLVQ IHVDTMKVIN DPIHGHIELH PLLVRIIDTP QFQRLRYIKQ LGGGYYVFPG A SHNRFEHS LGVGYLAGCL VHALGEKQPE LQISERDVLC VQIAGLCHDL GHGPFSHMFD GRFIPLARPE VKWTHEQGSV MM FEHLINS NGIKPVMEQY GLIPEEDICF IKEQIVGPLE SPVEDSLWPY KGRPENKSFL YEIVSNKRNG IDVDKWDYFA RDC HHLGIQ NNFDYKRFIK FARVCEVDNE LRICARDKEV GNLYDMFHTR NSLHRRAYQH KVGNIIDTMI TDAFLKADDY IEIT GAGGK KYRISTAIDD MEAYTKLTDN IFLEILYSTD PKLKDAREIL KQIEYRNLFK YVGETQPTGQ IKIKREDYES LPKEV ASAK PKVLLDVKLK AEDFIVDVIN MDYGMQEKNP IDHVSFYCKT APNRAIRITK NQVSQLLPEK FAEQLIRVYC KKVDRK SLY AARQYFVQWC ADRNFTKPQD GDVIAPLITP QKKEWNDSTS VQNPTRLREA SKSRVQLFKD DPM UniProtKB: Deoxynucleoside triphosphate triphosphohydrolase SAMHD1 |
-Macromolecule #2: 2'-deoxyguanosine-5'-O-(1-thiotriphosphate)
| Macromolecule | Name: 2'-deoxyguanosine-5'-O-(1-thiotriphosphate) / type: ligand / ID: 2 / Number of copies: 12 / Formula: T8T |
|---|---|
| Molecular weight | Theoretical: 523.247 Da |
| Chemical component information | 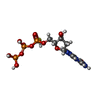 ChemComp-T8T: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.6 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 200 | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Number grids imaged: 1 / Number real images: 3222 / Average electron dose: 57.36 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.2 µm / Nominal defocus min: 0.6 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)