+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Post-fusion ectodomain of HSV-1 gB in complex with BMPC-23 Fab | |||||||||
 Map data Map data | HSV-1 gB in complex with BMPC23 Fab | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | glycoprotein / fusogen / antibody / ADCC / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationhost cell Golgi membrane / host cell endosome membrane / viral envelope / symbiont entry into host cell / virion attachment to host cell / host cell plasma membrane / virion membrane / identical protein binding / membrane Similarity search - Function | |||||||||
| Biological species |    Human alphaherpesvirus 1 strain KOS Human alphaherpesvirus 1 strain KOS | |||||||||
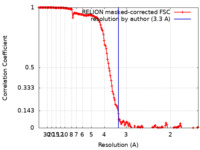
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Windsor IW / Kong SL / Garforth SJ / Almo SC / Harrison SC | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: J Clin Invest / Year: 2023 Journal: J Clin Invest / Year: 2023Title: A non-neutralizing glycoprotein B monoclonal antibody protects against herpes simplex virus disease in mice. Authors: Masayuki Kuraoka / Clare Burn Aschner / Ian W Windsor / Aakash Mahant Mahant / Scott J Garforth / Susan Luozheng Kong / Jacqueline M Achkar / Steven C Almo / Garnett Kelsoe / Betsy C Herold /  Abstract: There is an unmet need for monoclonal antibodies (mAbs) for prevention or as adjunctive treatment of herpes simplex virus (HSV) disease. Most vaccine and mAb efforts focus on neutralizing antibodies, ...There is an unmet need for monoclonal antibodies (mAbs) for prevention or as adjunctive treatment of herpes simplex virus (HSV) disease. Most vaccine and mAb efforts focus on neutralizing antibodies, but for HSV this strategy has proven ineffective. Preclinical studies with a candidate HSV vaccine strain, ΔgD-2, demonstrated that non-neutralizing antibodies that activate Fcγ receptors (FcγRs) to mediate antibody-dependent cellular cytotoxicity (ADCC) provide active and passive protection against HSV-1 and HSV-2. We hypothesized that this vaccine provides a tool to identify and characterize protective mAbs. We isolated HSV-specific mAbs from germinal center and memory B cells and bone marrow plasmacytes of ΔgD-2-vaccinated mice and evaluated these mAbs for binding, neutralizing, and FcγR-activating activity and for protective efficacy in mice. The most potent protective mAb, BMPC-23, was not neutralizing but activated murine FcγRIV, a biomarker of ADCC. The cryo-electron microscopic structure of the Fab-glycoprotein B (gB) assembly identified domain IV of gB as the epitope. A single dose of BMPC-23 administered 24 hours before or after viral challenge provided significant protection when configured as mouse IgG2c and protected mice expressing human FcγRIII when engineered as a human IgG1. These results highlight the importance of FcR-activating antibodies in protecting against HSV. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26520.map.gz emd_26520.map.gz | 315.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26520-v30.xml emd-26520-v30.xml emd-26520.xml emd-26520.xml | 16 KB 16 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_26520_fsc.xml emd_26520_fsc.xml | 15.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_26520.png emd_26520.png | 59.8 KB | ||
| Filedesc metadata |  emd-26520.cif.gz emd-26520.cif.gz | 5.7 KB | ||
| Others |  emd_26520_half_map_1.map.gz emd_26520_half_map_1.map.gz emd_26520_half_map_2.map.gz emd_26520_half_map_2.map.gz | 275.8 MB 275.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26520 http://ftp.pdbj.org/pub/emdb/structures/EMD-26520 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26520 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26520 | HTTPS FTP |
-Related structure data
| Related structure data |  7uhzMC  7ui0C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_26520.map.gz / Format: CCP4 / Size: 343 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26520.map.gz / Format: CCP4 / Size: 343 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | HSV-1 gB in complex with BMPC23 Fab | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.825 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: half map 2
| File | emd_26520_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 1
| File | emd_26520_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Trimeric HSV-1 gB in complex with three BCMP-23 Fabs
| Entire | Name: Trimeric HSV-1 gB in complex with three BCMP-23 Fabs |
|---|---|
| Components |
|
-Supramolecule #1: Trimeric HSV-1 gB in complex with three BCMP-23 Fabs
| Supramolecule | Name: Trimeric HSV-1 gB in complex with three BCMP-23 Fabs / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: BMPC-23 Fab Heavy chain
| Macromolecule | Name: BMPC-23 Fab Heavy chain / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 13.425734 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVQLQQSGPE LVKPGASVKL SCKASGYSFT TYDINWVKER PGQGLEWIGW IYPREGSTNY NEKFRGKATL TADTSSSTAY MELHSLTSE DSAVYFCATY GSSRYYTMDY WGQGTSVTVS SA |
-Macromolecule #2: BMPC-23 Fab Light chain
| Macromolecule | Name: BMPC-23 Fab Light chain / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 12.072557 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DIVLTQSPTS LAVSLGQRAT ISCRASESVD NFGISFMNWF QQKPGQPPKL LIYAASNLGS GVPARFSGSG SGTDFSLNIH PMEDDDTAM YFCQQSKEVP LTFGAGTKLE LKR |
-Macromolecule #3: Envelope glycoprotein B
| Macromolecule | Name: Envelope glycoprotein B / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human alphaherpesvirus 1 strain KOS / Strain: KOS Human alphaherpesvirus 1 strain KOS / Strain: KOS |
| Molecular weight | Theoretical: 71.668188 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: DIKAENTDAN FYVCPPPTGA TVVQFEQPRR CPTRPEGQNY TEGIAVVFKE NIAPYKFKAT MYYKDVTVSQ VWFGHRYSQF MGIFEDRAP VPFEEVIDKI NAKGVCRSTA KYVRNNLETT AFHRDDHETD MELKPANAAT RTSRGWHTTD LKYNPSRVEA F HRYGTTVN ...String: DIKAENTDAN FYVCPPPTGA TVVQFEQPRR CPTRPEGQNY TEGIAVVFKE NIAPYKFKAT MYYKDVTVSQ VWFGHRYSQF MGIFEDRAP VPFEEVIDKI NAKGVCRSTA KYVRNNLETT AFHRDDHETD MELKPANAAT RTSRGWHTTD LKYNPSRVEA F HRYGTTVN CIVEEVDARS VYPYDEFVLA TGDFVYMSPF YGYREGSHTE HTTYAADRFK QVDGFYARDL TTKARATAPT TR NLLTTPK FTVAWDWVPK RPSVCTMTKW QEVDEMLRSE YGGSFRFSSD AISTTFTTNL TEYPLSRVDL GDCIGKDARD AMD RIFARR YNATHIKVGQ PQYYQANGGF LIAYQPLLSN TLAELYVREH LREQSRKPPN PTPPPPGASA NASVERIKTT SSIE FARLQ FTYNHIQRHV NDMLGRVAIA WCELQNHELT LWNEARKLNP NAIASVTVGR RVSARMLGDV MAVSTCVPVA ADNVI VQNS MRISSRPGAC YSRPLVSFRY EDQGPLVEGQ LGENNELRLT RDAIEPCTVG HRRYFTFGGG YVYFEEYAYS HQLSRA DIT TVSTFIDLNI TMLEDHEFVP LEVYTRHEIK DSGLLDYTEV QRRNQLHDLR FADIDTVIHA DANAA UniProtKB: Envelope glycoprotein B |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 55.8 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.6 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)